3Q7T
 
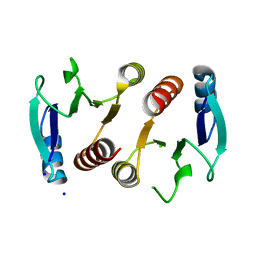 | | 2.15A resolution structure (I41 Form) of the ChxR receiver domain from Chlamydia trachomatis | | Descriptor: | SODIUM ION, Transcriptional regulatory protein | | Authors: | Hickey, J, Lovell, S, Battaile, K.P, Hu, L, Middaugh, C.R, Hefty, P.S. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The atypical response regulator protein ChxR has structural characteristics and dimer interface interactions that are unique within the OmpR/PhoB subfamily.
J.Biol.Chem., 286, 2011
|
|
3GG8
 
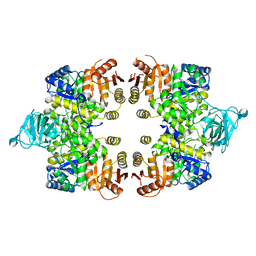 | | Crystal structure of the Toxoplasma gondii Pyruvate Kinase N terminal truncated | | Descriptor: | GLYCEROL, Pyruvate kinase, SULFATE ION | | Authors: | Wernimont, A.K, Lew, J, Allali-Hassani, A, Vedadi, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hills, T, Schapira, M, Hui, R, Pizarro, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of Toxoplasma gondii pyruvate kinase 1.
Plos One, 5, 2010
|
|
3QCU
 
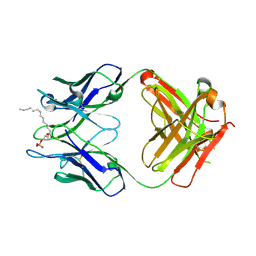 | | Crystal structure of the LT3015 antibody Fab fragment in complex with lysophosphatidic acid (14:0) | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, LT3015 antibody Fab fragment, heavy chain, ... | | Authors: | Fleming, J.K, Wojciak, J.M, Campbell, M.-A, Huxford, T. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Biochemical and structural characterization of lysophosphatidic Acid binding by a humanized monoclonal antibody.
J.Mol.Biol., 408, 2011
|
|
3GIC
 
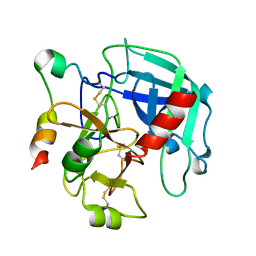 | | Structure of thrombin mutant delta(146-149e) in the free form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombin heavy chain, Thrombin light chain | | Authors: | Bah, A, Carrell, C.J, Chen, Z, Gandhi, P.S, Di Cera, E. | | Deposit date: | 2009-03-05 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Stabilization of the E* form turns thrombin into an anticoagulant.
J.Biol.Chem., 284, 2009
|
|
4JD5
 
 | | Crystal Structure of Benzoylformate Decarboxylase Mutant L403E | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Novak, W.R.P, Andrews, F.H, Tom, A.R, Gunderman, P.R, McLeish, M.J. | | Deposit date: | 2013-02-23 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A bulky hydrophobic residue is not required to maintain the v-conformation of enzyme-bound thiamin diphosphate.
Biochemistry, 52, 2013
|
|
2QFK
 
 | | X-ray Crystal Structure Analysis of the Binding Site in the Ferric and Oxyferrous Forms of the Recombinant Heme Dehaloperoxidase Cloned from Amphitrite ornata | | Descriptor: | AMMONIUM ION, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | de Serrano, V.S, Chen, Z, Davis, M.F, Franzen, S. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray crystal structural analysis of the binding site in the ferric and
oxyferrous forms of the recombinant heme dehaloperoxidase cloned from Amphitrite ornata
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4JG0
 
 | | Structure of phosphoserine/threonine (pSTAb) scaffold bound to pSer peptide | | Descriptor: | Fab heavy chain, Fab light chain, PROPANOIC ACID, ... | | Authors: | Koerber, J.T, Thomsen, N.D, Hannigan, B.T, Degrado, W.F, Wells, J.A. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Nature-inspired design of motif-specific antibody scaffolds.
Nat.Biotechnol., 31, 2013
|
|
3GCQ
 
 | | Human P38 MAP kinase in complex with RL45 | | Descriptor: | 1-{4-[(6-aminoquinazolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(3-methylphenyl)-1H-pyrazol-5-yl]urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-02-22 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of a fluorescent-tagged kinase assay system for the detection and characterization of allosteric kinase inhibitors.
J.Am.Chem.Soc., 131, 2009
|
|
3GIE
 
 | | Crystal structure of DesKC_H188E in complex with AMP-PCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Sensor histidine kinase desK | | Authors: | Trajtenberg, F, Albanesi, D, Alzari, P.M, Buschiazzo, A, de Mendoza, D. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural plasticity and catalysis regulation of a thermosensor histidine kinase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2QQM
 
 | | Crystal Structure of the a2b1b2 Domains from Human Neuropilin-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Appleton, B.A, Wiesmann, C. | | Deposit date: | 2007-07-26 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of neuropilin/antibody complexes provide insights into semaphorin and VEGF binding
Embo J., 26, 2007
|
|
4JEG
 
 | | Crystal Structure of Monobody CS1/SHP2 C-SH2 Domain Complex | | Descriptor: | Monobody CS1, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sha, F, Koide, S. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-28 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissection of the BCR-ABL signaling network using highly specific monobody inhibitors to the SHP2 SH2 domains.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3GF3
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with glutaconyl-coA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-02-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3QBP
 
 | |
3GG3
 
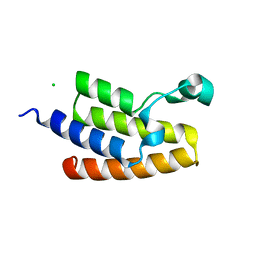 | | Crystal Structure of the Bromodomain of Human PCAF | | Descriptor: | CHLORIDE ION, Histone acetyltransferase PCAF | | Authors: | Filippakopoulos, P, Keates, T, Picaud, S, Rehana, K, Fedorov, O, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3GJC
 
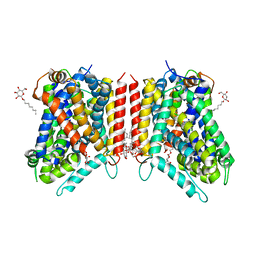 | | Crystal Structure of the E290S mutant of LeuT with bound OG | | Descriptor: | LEUCINE, SODIUM ION, Transporter, ... | | Authors: | Winther, A.M.L, Quick, M, Javitch, J.A, Nissen, P. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3QLR
 
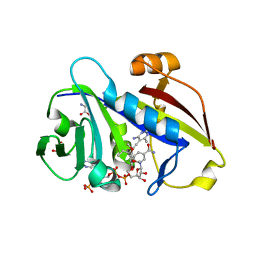 | | Candida albicans dihydrofolate reductase complexed with NADPH and 6-methyl-5-[(3R)-3-(3,4,5-trimethoxyphenyl)pent-1-yn-1-yl]pyrimidine-2,4-diamine (UCP112A) | | Descriptor: | 6-methyl-5-[(3R)-3-(3,4,5-trimethoxyphenyl)pent-1-yn-1-yl]pyrimidine-2,4-diamine, GLYCEROL, GLYCINE, ... | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
4JN9
 
 | | Crystal structure of the DepH | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DepH, ... | | Authors: | Li, J, Wang, C, Zhang, Z.M, Zhou, J.H, Cheng, E. | | Deposit date: | 2013-03-14 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of an NADP+-independent dithiol oxidase in FK228 biosynthesis.
Sci Rep, 4, 2014
|
|
3GK1
 
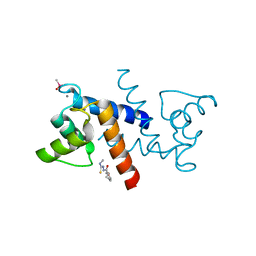 | | X-ray structure of bovine SBi132,Ca(2+)-S100B | | Descriptor: | 2-[(5-hex-1-yn-1-ylfuran-2-yl)carbonyl]-N-methylhydrazinecarbothioamide, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
3QF9
 
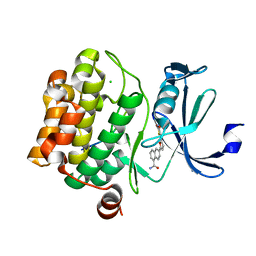 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a furan-thiazolidinedione ligand | | Descriptor: | 6-{5-[(Z)-(2,4-dioxo-1,3-thiazolidin-5-ylidene)methyl]furan-2-yl}-N-{3-[(4-ethylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl}naphthalene-1-carboxamide, CHLORIDE ION, Proto-oncogene serine/threonine-protein kinase pim-1, ... | | Authors: | Filippakopoulos, P, Bullock, A.N, Fedorov, O, Miduturu, C.V, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Grey, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-21 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a furan-thiazolidinedione ligand
To be Published
|
|
2QVU
 
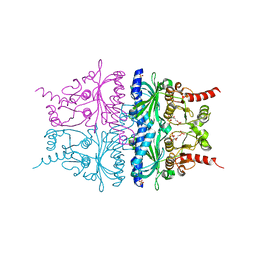 | | Porcine Liver Fructose-1,6-bisphosphatase cocrystallized with Fru-2,6-P2 and Mg2+, I(T)-state | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase 1, MAGNESIUM ION, ... | | Authors: | Hines, J.K, Chen, X, Nix, J.C, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of mammalian and bacterial fructose-1,6-bisphosphatase reveal the basis for synergism in AMP/fructose 2,6-bisphosphate inhibition
J.Biol.Chem., 282, 2007
|
|
3GKF
 
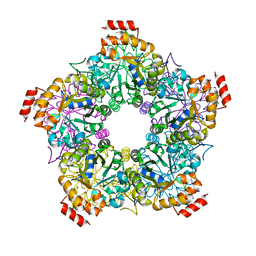 | | Crystal Structure of E. coli LsrF | | Descriptor: | Aldolase lsrF | | Authors: | Miller, S.T, Diaz, Z.C. | | Deposit date: | 2009-03-10 | | Release date: | 2009-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of the Escherichia coli autoinducer-2 processing protein LsrF.
Plos One, 4, 2009
|
|
3Q60
 
 | |
4JPO
 
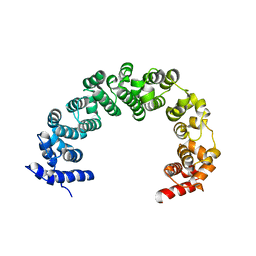 | | 5A resolution structure of Proteasome Assembly Chaperone Hsm3 in complex with a C-terminal fragment of Rpt1 | | Descriptor: | 26S protease regulatory subunit 7 homolog, DNA mismatch repair protein HSM3 | | Authors: | Lovell, S, Battaile, K.P, Singh, R, Roelofs, J. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Reconfiguration of the proteasome during chaperone-mediated assembly.
Nature, 497, 2013
|
|
3GNQ
 
 | |
3QIJ
 
 | | Primitive-monoclinic crystal structure of the FERM domain of protein 4.1R | | Descriptor: | Protein 4.1, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Zhong, N, Tong, Y, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-27 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Primitive-monoclinic crystal structure of the FERM domain of protein 4.1R
to be published
|
|
