1Q18
 
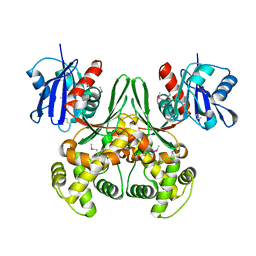 | | Crystal structure of E.coli glucokinase (Glk) | | Descriptor: | Glucokinase | | Authors: | Lunin, V.V, Li, Y, Schrag, J.D, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-07-18 | | Release date: | 2004-07-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose.
J.Bacteriol., 186, 2004
|
|
1PW6
 
 | | Low Micromolar Small Molecule Inhibitor of IL-2 | | Descriptor: | 2-CYCLOHEXYL-N-(2-{4-[5-(2,3-DICHLORO-PHENYL)-2H-PYRAZOL-3-YL]-PIPERIDIN-1-YL}-2-OXO-ETHYL)-2-GUANIDINO-ACETAMIDE, Interleukin-2, SULFATE ION | | Authors: | Thanos, C.D, Randal, M, Wells, J.A. | | Deposit date: | 2003-06-30 | | Release date: | 2004-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Potent small-molecule binding to a dynamic hot spot on IL-2.
J.Am.Chem.Soc., 125, 2003
|
|
1PY2
 
 | | Structure of a 60 nM Small Molecule Bound to a Hot Spot on IL-2 | | Descriptor: | 5-[2,3-DICHLORO-4-(5-{1-[2-(2-GUANIDINO-4-METHYL-PENTANOYLAMINO)-ACETYL]-PIPERIDIN-4-YL}-1-METHYL-1H-PYRAZOL-3-YL)-PHENOXYMETHYL]-FURAN-2-CARBOXYLIC ACID, Interleukin-2, ZINC ION | | Authors: | Thanos, C.D, Randal, M, Wells, J.A. | | Deposit date: | 2003-07-07 | | Release date: | 2004-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Potent small-molecule binding to a dynamic hot spot on IL-2.
J.Am.Chem.Soc., 125, 2003
|
|
1SZ2
 
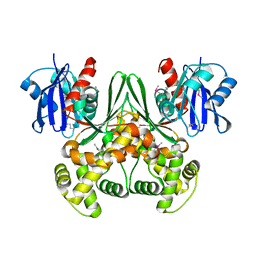 | | Crystal structure of E. coli glucokinase in complex with glucose | | Descriptor: | Glucokinase, beta-D-glucopyranose | | Authors: | Lunin, V.V, Li, Y, Schrag, J.D, Iannuzzi, P, Matte, A, Cygler, M. | | Deposit date: | 2004-04-02 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose
J.Bacteriol., 186, 2004
|
|
3CDO
 
 | |
3CH3
 
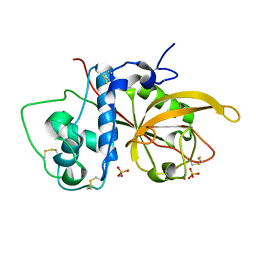 | | Crystal Structure Analysis of SERA5E from plasmodium falciparum | | Descriptor: | DIHYDROGENPHOSPHATE ION, POTASSIUM ION, Serine-repeat antigen protein | | Authors: | Smith, B.J, Malby, R.L, Colman, P.M, Clarke, O.B. | | Deposit date: | 2008-03-07 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Insights into the Protease-like Antigen Plasmodium falciparum SERA5 and Its Noncanonical Active-Site Serine
J.Mol.Biol., 392, 2009
|
|
3D7P
 
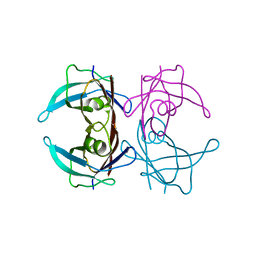 | | Crystal structure of human Transthyretin (TTR) at pH 4.0 | | Descriptor: | Transthyretin | | Authors: | Palaninathan, S.K, Mohamedmohaideen, N.N, Snee, W.C, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2008-05-21 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural insight into pH-induced conformational changes within the native human transthyretin tetramer.
J.Mol.Biol., 382, 2008
|
|
5F21
 
 | | human CD38 in complex with nanobody MU375 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Hao, Q, Zhang, H. | | Deposit date: | 2015-12-01 | | Release date: | 2016-06-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
5F1K
 
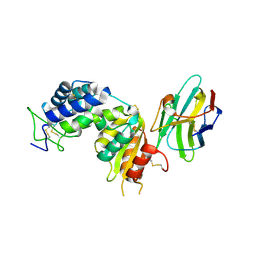 | | human CD38 in complex with nanobody MU1053 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, nanobody MU1053 | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
5F1O
 
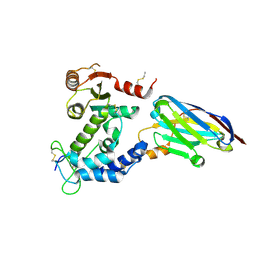 | | human CD38 in complex with nanobody MU551 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, nanobody MU551 | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
8W0Q
 
 | | Pembrolizumab CDR-H3 Loop Mimic | | Descriptor: | Pembrolizumab CDR-H3 Loop Mimic | | Authors: | Feig, M, Roche, S.P. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | De Novo Synthesis and Structural Elucidation of CDR-H3 Loop Mimics.
Acs Chem.Biol., 19, 2024
|
|
1MZ4
 
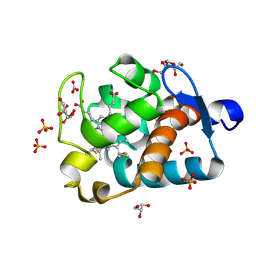 | | Crystal Structure of Cytochrome c550 from Thermosynechococcus elongatus | | Descriptor: | BICARBONATE ION, GLYCEROL, HEME C, ... | | Authors: | Kerfeld, C.A, Sawaya, M.R, Bottin, H, Tran, K.T, Sugiura, M, Kirilovsky, D, Krogmann, D, Yeates, T.O, Boussac, A. | | Deposit date: | 2002-10-05 | | Release date: | 2003-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and EPR characterization of the soluble form of cytochrome c-550 and of the psbV2 gene product from the cyanobacterium Thermosynechococcus elongatus.
Plant Cell.Physiol., 44, 2003
|
|
5DMQ
 
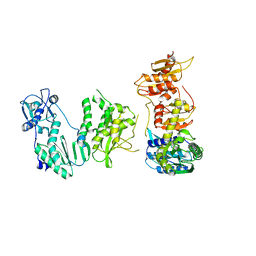 | |
9EOQ
 
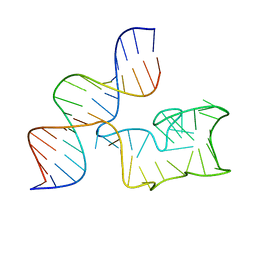 | | Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 and TBA | | Descriptor: | DNA (42-MER), DNA (5'-D(P*AP*TP*AP*TP*AP*GP*CP*GP*TP*GP*GP*AP*AP*GP*T)-3') | | Authors: | Ali, K, Georg, K, Volodymyr, M, Johanna, G, Maximilian, N.H, Lukas, K, Simone, C, Hendrik, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Designing Rigid DNA Origami Templates for Molecular Visualization Using Cryo-EM.
Nano Lett., 24, 2024
|
|
4EZV
 
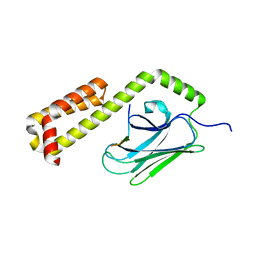 | |
8VCZ
 
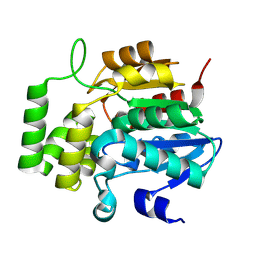 | |
8VD3
 
 | |
8VD1
 
 | | Crystal Structure of KAI2 from Oryza sativa with MPD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Probable esterase D14L | | Authors: | Gilio, A.K, Shabek, N, Guercio, A.M, Pawlak, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structural insights into rice KAI2 receptor provide functional implications for perception and signal transduction.
J.Biol.Chem., 300, 2024
|
|
5G5D
 
 | | Crystal Structure of the CohScaC2-XDocCipA type II complex from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CELLULOSOMAL-SCAFFOLDING PROTEIN A, CELLULOSOME ANCHORING PROTEIN COHESIN REGION | | Authors: | Carvalho, A.L, A Bras, J.L, Najmudin, S.H, Pinheiro, B.A, Fontes, C.M.G.A. | | Deposit date: | 2016-05-23 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
4EZU
 
 | |
3LGU
 
 | | Y162A mutant of the DegS-deltaPDZ protease | | Descriptor: | Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
3LGT
 
 | |
8FAV
 
 | |
6W50
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR METHYL ((10R,14S)- 14-(4-(3-CHLORO-2,6-DIFLUOROPHENYL)-6-OXO-3,6-DIHYDRO- 1(2H)-PYRIDINYL)-10-METHYL-9-OXO-8,16- DIAZATRICYCLO[13.3.1.0~2,7~]NONADECA-1(19),2,4,6,15,17- HEXAEN-5-YL)CARBAMATE | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2020-03-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a High Affinity, Orally Bioavailable Macrocyclic FXIa Inhibitor with Antithrombotic Activity in Preclinical Species.
J.Med.Chem., 63, 2020
|
|
2IVY
 
 | | Crystal structure of hypothetical protein sso1404 from Sulfolobus solfataricus P2 | | Descriptor: | HYPOTHETICAL PROTEIN SSO1404 | | Authors: | Yan, X, Carter, L.G, Dorward, M, Liu, H, McMahon, S.A, Oke, M, Powers, H, White, M.F, Naismith, J.H. | | Deposit date: | 2006-06-22 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
