3JWO
 
 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D Heavy CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Huang, C.C, Kwon, Y.D, Zhou, T, Robinson, J.E, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JWD
 
 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D HEAVY CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Ban, Y.A, Chen, L, Huang, C.C, Kong, L, Kwon, Y.D, Stuckey, J, Zhou, T, Robinson, J.E, Schief, W.R, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KDK
 
 | |
3KDG
 
 | |
1C52
 
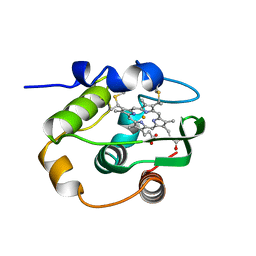 | | THERMUS THERMOPHILUS CYTOCHROME-C552: A NEW HIGHLY THERMOSTABLE CYTOCHROME-C STRUCTURE OBTAINED BY MAD PHASING | | Descriptor: | CYTOCHROME-C552, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Than, M.E, Hof, P, Huber, R, Bourenkov, G.P, Bartunik, H.D, Buse, G, Soulimane, T. | | Deposit date: | 1997-06-23 | | Release date: | 1998-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Thermus thermophilus cytochrome-c552: A new highly thermostable cytochrome-c structure obtained by MAD phasing.
J.Mol.Biol., 271, 1997
|
|
1ADJ
 
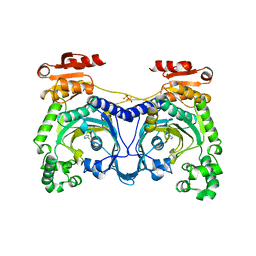 | | HISTIDYL-TRNA SYNTHETASE IN COMPLEX WITH HISTIDINE | | Descriptor: | HISTIDINE, HISTIDYL-TRNA SYNTHETASE, SULFATE ION | | Authors: | Cusack, S, Aberg, A. | | Deposit date: | 1997-02-18 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure analysis of the activation of histidine by Thermus thermophilus histidyl-tRNA synthetase.
Biochemistry, 36, 1997
|
|
3JWN
 
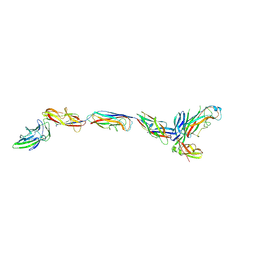 | | Complex of FimC, FimF, FimG and FimH | | Descriptor: | Chaperone protein fimC, FimH protein, GLYCEROL, ... | | Authors: | Le Trong, I, Aprikian, P, Stenkamp, R.E, Sokurenko, E.V. | | Deposit date: | 2009-09-18 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like beta sheet twisting.
Cell(Cambridge,Mass.), 141, 2010
|
|
3K1F
 
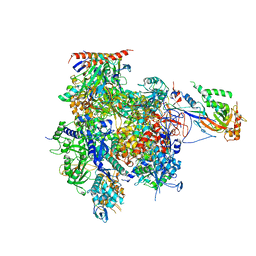 | | Crystal structure of RNA Polymerase II in complex with TFIIB | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Kostrewa, D, Zeller, M.E, Armache, K.-J, Seizl, M, Leike, K, Thomm, M, Cramer, P. | | Deposit date: | 2009-09-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | RNA polymerase II-TFIIB structure and mechanism of transcription initiation.
Nature, 462, 2009
|
|
1B43
 
 | | FEN-1 FROM P. FURIOSUS | | Descriptor: | PROTEIN (FEN-1) | | Authors: | Hosfield, D.J, Mol, C.D, Shen, B, Tainer, J.A. | | Deposit date: | 1999-01-05 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity.
Cell(Cambridge,Mass.), 95, 1998
|
|
1CZ1
 
 | | EXO-B-(1,3)-GLUCANASE FROM CANDIDA ALBICANS AT 1.85 A RESOLUTION | | Descriptor: | PROTEIN (EXO-B-(1,3)-GLUCANASE) | | Authors: | Cutfield, S.M, Davies, G.J, Murshudov, G, Anderson, B.F, Moody, P.C.E, Sullivan, P.A, Cutfield, J.F. | | Deposit date: | 1999-09-01 | | Release date: | 2000-01-03 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of the exo-beta-(1,3)-glucanase from Candida albicans in native and bound forms: relationship between a pocket and groove in family 5 glycosyl hydrolases.
J.Mol.Biol., 294, 1999
|
|
5AIN
 
 | | Varenicline Interactions at the 5HT3 Receptor Ligand Binding Site are Revealed by 5HTBP | | Descriptor: | SOLUBLE ACETYLCHOLINE RECEPTOR, VARENICLINE | | Authors: | Price, K.L, Lillestol, R, Ulens, C, Lummis, S.C. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Varenicline Interactions at the 5-Ht3 Receptor Ligand Binding Site are Revealed by 5-Htbp.
Acs Chem Neurosci, 6, 2015
|
|
329D
 
 | |
5YD0
 
 | | Crystal structure of Schlafen 13 (SLFN13) N'-domain | | Descriptor: | Schlafen 8, ZINC ION | | Authors: | Yang, J.-Y, Gao, S. | | Deposit date: | 2017-09-09 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.182 Å) | | Cite: | Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control
Nat Commun, 9, 2018
|
|
5Y6J
 
 | |
5C0N
 
 | |
5WCE
 
 | | Caulobacter crescentus pol III beta | | Descriptor: | DNA polymerase III subunit beta | | Authors: | Oakley, A.J. | | Deposit date: | 2017-06-30 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Pol III beta from Caulobacter crescentus
To Be Published
|
|
4Z9P
 
 | | Crystal structure of Ebola virus nucleoprotein core domain at 1.8A resolution | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Dong, S.S, Yang, P, Li, G.B, Liu, B.C, Yang, C, Rao, Z.H. | | Deposit date: | 2015-04-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Insight into the Ebola virus nucleocapsid assembly mechanism: crystal structure of Ebola virus nucleoprotein core domain at 1.8 A resolution.
Protein Cell, 6, 2015
|
|
2ZGV
 
 | | Crystal Structure of human phosphoglycerate kinase bound to D-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoglycerate kinase 1 | | Authors: | Arold, S.T, Gondeau, C, Lionne, C, Chaloin, L. | | Deposit date: | 2008-01-26 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the lack of enantioselectivity of human 3-phosphoglycerate kinase
Nucleic Acids Res., 36, 2008
|
|
5WTA
 
 | | Crystal Structure of Staphylococcus aureus SdrE apo form | | Descriptor: | Serine-aspartate repeat-containing protein E | | Authors: | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | Deposit date: | 2016-12-10 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
3C39
 
 | | Crystal Structure of human phosphoglycerate kinase bound to 3-phosphoglycerate | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Phosphoglycerate kinase 1 | | Authors: | Arold, S.T, Gondeau, C, Lionne, C, Chaloin, L. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis for the lack of enantioselectivity of human 3-phosphoglycerate kinase
Nucleic Acids Res., 36, 2008
|
|
5CDH
 
 | |
4IRE
 
 | | Crystal structure of GLIC with mutations at the loop C region | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ACETATE ION, OXALATE ION, ... | | Authors: | Chen, Q, Pan, J, Liang, Y.H, Xu, Y, Tang, P. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Signal transduction pathways in the pentameric ligand-gated ion channels.
Plos One, 8, 2013
|
|
3BEH
 
 | |
3BDL
 
 | | Crystal structure of a truncated human Tudor-SN | | Descriptor: | CITRIC ACID, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Li, C.L. | | Deposit date: | 2007-11-15 | | Release date: | 2008-08-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing.
Nucleic Acids Res., 36, 2008
|
|
5WTB
 
 | | Complex Structure of Staphylococcus aureus SdrE with human complement factor H | | Descriptor: | Peptide from Complement factor H, Serine-aspartate repeat-containing protein E | | Authors: | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | Deposit date: | 2016-12-10 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
