4KTT
 
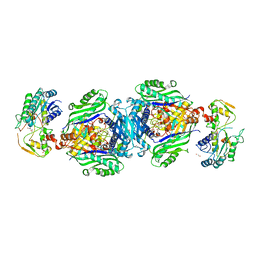 | | Structural insights of MAT enzymes: MATa2b complexed with SAM | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methionine adenosyltransferase 2 subunit beta, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2013-05-21 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure and function study of the complex that synthesizes S-adenosylmethionine.
IUCrJ, 1, 2014
|
|
5UMH
 
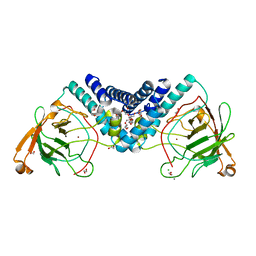 | |
7XMG
 
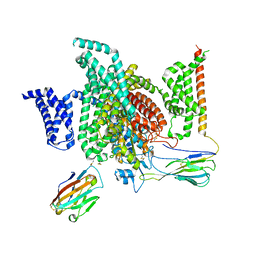 | | Cryo-EM structure of human NaV1.7/beta1/beta2-TCN-1752 | | Descriptor: | (1~{Z})-~{N}-[2-methyl-3-[(~{E})-[6-[4-[[4-(trifluoromethyloxy)phenyl]methoxy]piperidin-1-yl]-1~{H}-1,3,5-triazin-2-ylidene]amino]phenyl]ethanimidic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5TYR
 
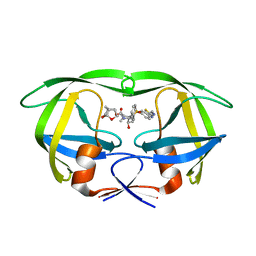 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-121 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel central nervous system-penetrating protease inhibitor overcomes human immunodeficiency virus 1 resistance with unprecedented aM to pM potency.
Elife, 6, 2017
|
|
5TYS
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | A novel central nervous system-penetrating protease inhibitor overcomes human immunodeficiency virus 1 resistance with unprecedented aM to pM potency.
Elife, 6, 2017
|
|
7FIQ
 
 | | The crystal structure of mannose-bound beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | Descriptor: | Beta-1,2-mannobiose phosphorylase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
6HMH
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-glucosamine) and alpha-1,2-mannobiose | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},6~{S})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Lu, D, Zhu, S, Bernardo-Seisdedos, G, Millet, O, Zhang, Y, Sollogoub, M, Jimenez-Barbero, J, Davies, G.J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | From 1,4-Disaccharide to 1,3-Glycosyl Carbasugar: Synthesis of a Bespoke Inhibitor of Family GH99 Endo-alpha-mannosidase.
Org.Lett., 20, 2018
|
|
4JBE
 
 | | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, ... | | Authors: | Minasov, G, Filippova, E.V, Halavaty, A, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis.
TO BE PUBLISHED
|
|
6Q3R
 
 | | ASPERGILLUS ACULEATUS GALACTANASE | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ACETATE ION, ... | | Authors: | Muderspach, S.J, Torpenholt, S, Lo Leggio, L, Poulsen, J.C.N. | | Deposit date: | 2018-12-04 | | Release date: | 2019-06-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of Aspergillus aculeatus beta-1,4-galactanase in complex with galactobiose.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
3I4V
 
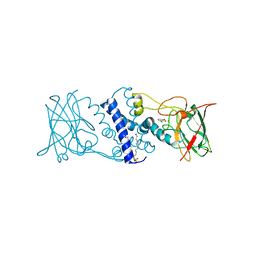 | | Crystal structure determination of catechol 1,2-dioxygenase from rhodococcus opacus 1CP in complex with 3-chlorocatechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 3-chlorobenzene-1,2-diol, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-07-03 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
3I51
 
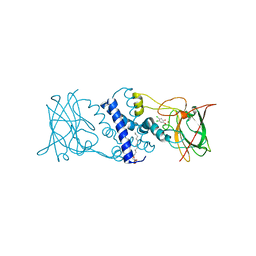 | | Crystal structure determination of Catechol 1,2-Dioxygenase from Rhodococcus opacus 1CP in complex with 4,5-dichlorocatechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 4,5-dichlorobenzene-1,2-diol, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Kolomytseva, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-07-03 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
6KDN
 
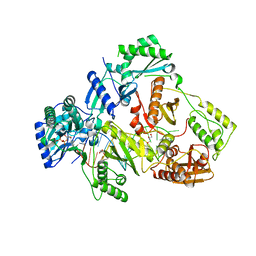 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
4NV4
 
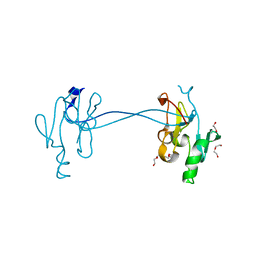 | | 1.8 Angstrom Crystal Structure of Signal Peptidase I from Bacillus anthracis. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Signal peptidase I, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-04 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of Signal Peptidase I from Bacillus anthracis.
TO BE PUBLISHED
|
|
3I4Y
 
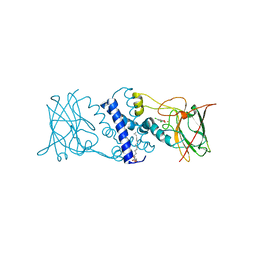 | | Crystal structure determination of Catechol 1,2-Dioxygenase from Rhodococcus opacus 1CP in complex with 3,5-dichlorocatechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 3,5-dichlorobenzene-1,2-diol, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Kolomytseva, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-07-03 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
5U6V
 
 | | X-ray crystal structure of 1,2,3-triazolobenzodiazepine in complex with BRD2(D2) | | Descriptor: | 1,2-ETHANEDIOL, 5-[7-(4-chlorophenyl)-1-methyl-6,7-dihydro-5H-[1,2,3]triazolo[1,5-d][1,4]benzodiazepin-9-yl]pyridin-2-amine, Bromodomain-containing protein 2 | | Authors: | Hatfaludi, T, Sharp, P.P, Garnier, J.-M, Burns, C.J, Czabotar, P.E. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.775 Å) | | Cite: | Design, Synthesis, and Biological Activity of 1,2,3-Triazolobenzodiazepine BET Bromodomain Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5DPX
 
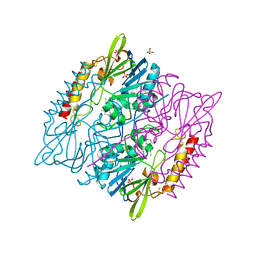 | | 1,2,4-Triazole-3-thione compounds as inhibitors of L1, di-zinc metallo-beta-lactamases. | | Descriptor: | 5-(2-methylphenyl)-3H-1,2,4-triazole-3-thione, Metallo-beta-lactamase L1 type 3, SULFATE ION, ... | | Authors: | Nauton, L, Garau, G, Khan, R, Dideberg, O. | | Deposit date: | 2015-09-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,2,4-Triazole-3-thione Compounds as Inhibitors of Dizinc Metallo-beta-lactamases.
ChemMedChem, 12, 2017
|
|
2WMV
 
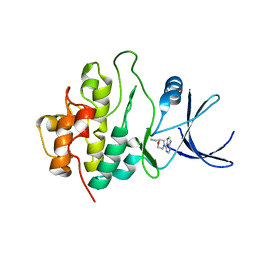 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1-[(2S)-4-(7H-PURIN-6-YL)MORPHOLIN-2-YL]METHANAMINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
6L9T
 
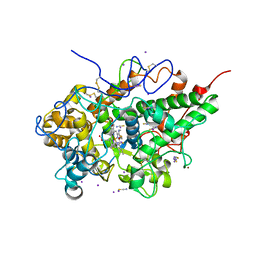 | | Crystal structure of the complex of bovine lactoperoxidase with OSCN at 1.89 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Viswanathan, V, Pandey, N, Singh, A, Sinha, M, Singh, R.P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the complex of bovine lactoperoxidase with OSCN at 1.89 A resolution
To Be Published
|
|
6MCS
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-003 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | Authors: | Bulut, H, Hayashi, H, Hattori, S.I, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-09-02 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.
Antimicrob.Agents Chemother., 63, 2019
|
|
6MNC
 
 | | CRYSTAL STRUCTURE OF HUMAN 17BETA-HYDROXYSTEROID DEHYDROGENASE TYPE 1 COMPLEXED WITH ESTRONE | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, DI(HYDROXYETHYL)ETHER, Estradiol 17-beta-dehydrogenase 1 | | Authors: | Li, T, Stephen, P, Zhu, D.W, Lin, S.X. | | Deposit date: | 2018-10-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of human 17 beta-hydroxysteroid dehydrogenase type 1 complexed with estrone and NADP+reveal the mechanism of substrate inhibition.
Febs J., 286, 2019
|
|
5XVK
 
 | | Crystal structure of mouse Nicotinamide N-methyltransferase (NNMT) bound with end product, 1-methyl Nicotinamide (MNA) | | Descriptor: | 3-carbamoyl-1-methylpyridin-1-ium, GLYCEROL, Nicotinamide N-methyltransferase, ... | | Authors: | Swaminathan, S, Birudukota, S, Thakur, M.K, Parveen, R, Kandan, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of monkey and mouse nicotinamide N-methyltransferase (NNMT) bound with end product, 1-methyl nicotinamide
Biochem. Biophys. Res. Commun., 491, 2017
|
|
6MNE
 
 | | CRYSTAL STRUCTURE OF HUMAN 17BETA-HYDROXYSTEROID DEHYDROGENASE TYPE 1 COMPLEXED WITH ESTRONE AND NADP+ | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, Estradiol 17-beta-dehydrogenase 1, GLYCEROL, ... | | Authors: | Li, T, Lin, S.X. | | Deposit date: | 2018-10-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of human 17 beta-hydroxysteroid dehydrogenase type 1 complexed with estrone and NADP+reveal the mechanism of substrate inhibition.
Febs J., 286, 2019
|
|
3HKP
 
 | | Crystal structure determination of Catechol 1,2-Dioxygenase from Rhodococcus opacus 1CP in complex with protocatechuate | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 3,4-DIHYDROXYBENZOIC ACID, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Kolomytseva, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-05-25 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
3OYT
 
 | | 1.84 Angstrom resolution crystal structure of 3-oxoacyl-(acyl carrier protein) synthase I (fabB) from Yersinia pestis CO92 | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] synthase I, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Peterson, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-23 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | 1.84 Angstrom resolution crystal structure of 3-oxoacyl-(acyl carrier protein) synthase I (fabB) from Yersinia pestis CO92
To be Published
|
|
6KDM
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:entecavir 5'-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
