1C6Y
 
 | |
1C70
 
 | |
1O95
 
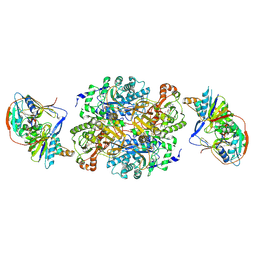 | | Ternary complex between trimethylamine dehydrogenase and electron transferring flavoprotein | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ELECTRON TRANSFER FLAVOPROTEIN ALPHA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
3MTI
 
 | |
1D15
 
 | | TERNARY INTERACTIONS OF SPERMINE WITH DNA: 4'-EPIADRIAMYCIN AND OTHER DNA: ANTHRACYCLINE COMPLEXES | | Descriptor: | 4'-EPIDOXORUBICIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), SPERMINE | | Authors: | Williams, L.D, Frederick, C.A, Ughetto, G, Rich, A. | | Deposit date: | 1990-07-03 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ternary interactions of spermine with DNA: 4'-epiadriamycin and other DNA: anthracycline complexes.
Nucleic Acids Res., 18, 1990
|
|
8TZX
 
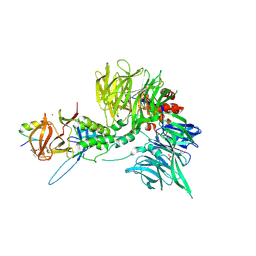 | | Ternary complex structure of Cereblon-DDB1 bound to WIZ(ZF7) and the molecular glue dWIZ-1 | | Descriptor: | (3S)-3-(5-{(1R)-1-[(2R)-1-ethylpiperidin-2-yl]ethoxy}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, DNA damage-binding protein 1, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-28 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Ternary complex structure of Cereblon-DDB1 bound to WIZ(ZF7) and the molecular glue dWIZ-1
Science, 2024
|
|
2JP7
 
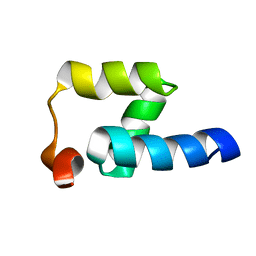 | | NMR structure of the Mex67 UBA domain | | Descriptor: | mRNA export factor MEX67 | | Authors: | Hobeika, M, Brockmann, C, Iglesias, N, Gwizdek, C, Neuhaus, D, Stutz, F, Stewart, M, Divita, G, Dargemont, C. | | Deposit date: | 2007-04-26 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Coordination of Hpr1 and ubiquitin binding by the UBA domain of the mRNA export factor Mex67
Mol.Cell.Biol., 18, 2007
|
|
2OKH
 
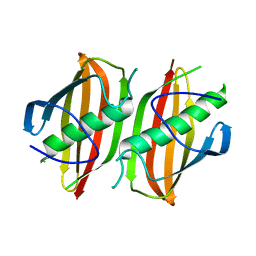 | | Crystal structure of dimeric form of PfFabZ in crystal form3 | | Descriptor: | Beta-hydroxyacyl-ACP dehydratase | | Authors: | Swarnamukhi, P.L, Sharma, S.K, Padala, P, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2007-01-16 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Packing and loop-structure variations in non-isomorphous crystals of FabZ from Plasmodium falciparum
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2OKI
 
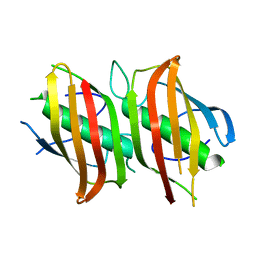 | | Crystal structure of dimeric form of PfFabZ in crystal form2 | | Descriptor: | Beta-hydroxyacyl-ACP dehydratase | | Authors: | Swarnamukhi, P.L, Sharma, S.K, Padala, P, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2007-01-17 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Packing and loop-structure variations in non-isomorphous crystals of FabZ from Plasmodium falciparum
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2QOM
 
 | | The crystal structure of the E.coli EspP autotransporter Beta-domain. | | Descriptor: | Serine protease espP | | Authors: | Barnard, T.J, Dautin, N, Lukacik, P, Bernstein, H.D, Buchanan, S.K. | | Deposit date: | 2007-07-20 | | Release date: | 2007-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Autotransporter structure reveals intra-barrel cleavage followed by conformational changes.
Nat.Struct.Mol.Biol., 14, 2007
|
|
3D3Q
 
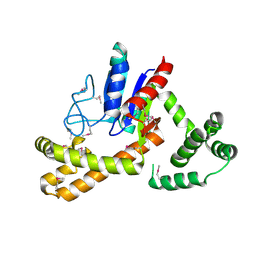 | | Crystal structure of tRNA delta(2)-isopentenylpyrophosphate transferase (SE0981) from Staphylococcus epidermidis. Northeast Structural Genomics Consortium target SeR100 | | Descriptor: | tRNA delta(2)-isopentenylpyrophosphate transferase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Mao, L, Xiao, R, Maglaqui, M, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of tRNA delta(2)-isopentenylpyrophosphate transferase (SE0981) from Staphylococcus epidermidis.
To be Published
|
|
1XKD
 
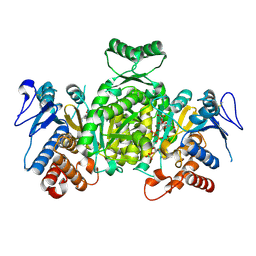 | | Ternary complex of Isocitrate dehydrogenase from the hyperthermophile Aeropyrum pernix | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Karlstrom, M, Stokke, R, Steen, I.H, Birkeland, N.-K, Ladenstein, R. | | Deposit date: | 2004-09-28 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Isocitrate dehydrogenase from the hyperthermophile Aeropyrum pernix: X-ray structure analysis of a ternary enzyme-substrate complex and thermal stability.
J.Mol.Biol., 345, 2005
|
|
1DY3
 
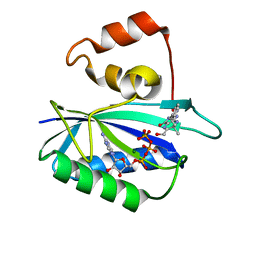 | | Ternary complex of 7,8-dihydro-6-hydroxymethylpterinpyrophosphokinase from Escherichia coli with ATP and a substrate analogue. | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 7,8-DIHYDRO-6-HYDROXYMETHYL-7-METHYL-7-[2-PHENYLETHYL]-PTERIN, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Stammers, D.K, Achari, A, Somers, D.O, Bryant, P.K, Rosemond, J, Scott, D.L, Champness, J.N. | | Deposit date: | 2000-01-21 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A X-Ray Structure of the Ternary Complex of 7,8-Dihydro-6-Hydroxymethylpterinpyrophosphokinase from Escherichia Coli with ATP and a Substrate Analogue
FEBS Lett., 456, 1999
|
|
1G9H
 
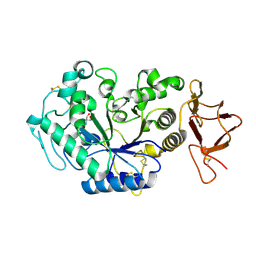 | | TERNARY COMPLEX BETWEEN PSYCHROPHILIC ALPHA-AMYLASE, COMII (PSEUDO TRI-SACCHARIDE FROM BAYER) AND TRIS (2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose, ALPHA-AMYLASE, ... | | Authors: | Aghajari, N, Roth, M, Haser, R. | | Deposit date: | 2000-11-23 | | Release date: | 2002-06-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic evidence of a transglycosylation reaction: ternary complexes of a
psychrophilic alpha-amylase.
Biochemistry, 41, 2002
|
|
1MA0
 
 | | Ternary complex of Human glutathione-dependent formaldehyde dehydrogenase with NAD+ and dodecanoic acid | | Descriptor: | Glutathione-dependent formaldehyde dehydrogenase, LAURIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sanghani, P.C, Robinson, H, Bosron, W.F, Hurley, T.D. | | Deposit date: | 2002-07-30 | | Release date: | 2002-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human glutathione-dependent formaldehyde dehydrogenase. Structures of apo, binary, and inhibitory ternary complexes.
Biochemistry, 41, 2002
|
|
2OH2
 
 | | Ternary Complex of Human DNA Polymerase | | Descriptor: | 5'-D(*GP*GP*G*GP*GP*AP*AP*GP*GP*AP*CP*CP*C)-3', 5'-D(*TP*T*CP*CP*AP*GP*GP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', DNA polymerase kappa, ... | | Authors: | Lone, S, Townson, S.A, Uljon, S.N, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2007-01-09 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Ternary complex of the catalytic core of human DNA polymerase Kappa with DNA and dTT
To be Published
|
|
7QWH
 
 | | X-ray structure of the adduct formed upon reaction of a vanadium hydroxyquinoline complex with RNase A | | Descriptor: | 2,2-bis($l^{1}-oxidanyl)-3-oxa-1$l^{4}-aza-2$l^{4}-vanadatricyclo[6.3.1.0^{4,12}]dodeca-1(12),4,6,8,10-pentaene, Ribonuclease pancreatic | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2022-01-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Interaction of VIVO-8-hydroxyquinoline species with RNase A: the effect of metal ligands in the protein adduct stabilization
Inorg Chem Front, 2023
|
|
4FH7
 
 | | Structure of DHP A in complex with 2,4,6-tribromophenol in 20% methanol | | Descriptor: | 2,4,6-TRIBROMOPHENOL, Dehaloperoxidase A, METHANOL, ... | | Authors: | de Serrano, V.S, Franzen, S. | | Deposit date: | 2012-06-05 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Kinetic Study of an Internal Substrate Binding Site in Dehaloperoxidase-Hemoglobin A from Amphitrite ornata.
Biochemistry, 52, 2013
|
|
4FH6
 
 | | Structure of DHP A in complex with 2,4,6-tribromophenol in 10% DMSO | | Descriptor: | 2,4,6-TRIBROMOPHENOL, DIMETHYL SULFOXIDE, Dehaloperoxidase A, ... | | Authors: | de Serrano, V.S, Franzen, S. | | Deposit date: | 2012-06-05 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural and Kinetic Study of an Internal Substrate Binding Site in Dehaloperoxidase-Hemoglobin A from Amphitrite ornata.
Biochemistry, 52, 2013
|
|
3UF0
 
 | | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase SDR | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy)
To be Published
|
|
6MCI
 
 | | Solution structure of 7SK stem-loop 1 | | Descriptor: | 7SK RNA | | Authors: | Pham, V.V, D'Souza, V.M. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | HIV-1 Tat interactions with cellular 7SK and viral TAR RNAs identifies dual structural mimicry.
Nat Commun, 9, 2018
|
|
5VLX
 
 | | Dehaloperoxidase B mutant F21W | | Descriptor: | Dehaloperoxidase B, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Carey, L.M, Ghiladi, R.A. | | Deposit date: | 2017-04-26 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective tuning of activity in a multifunctional enzyme as revealed in the F21W mutant of dehaloperoxidase B from Amphitrite ornata.
J. Biol. Inorg. Chem., 23, 2018
|
|
7M1K
 
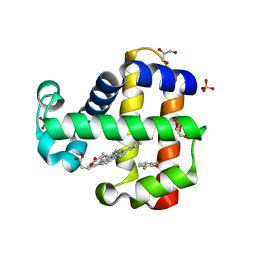 | | Crystal structure of dehaloperoxidase B in complex with 2,6-difluorophenol | | Descriptor: | 2,6-DIFLUOROPHENOL, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T. | | Deposit date: | 2021-03-13 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Bridging the functional gap between reactivity and inhibition in dehaloperoxidase B from Amphitrite ornata: Mechanistic and structural studies with 2,4- and 2,6-dihalophenols.
J.Inorg.Biochem., 236, 2022
|
|
7M1J
 
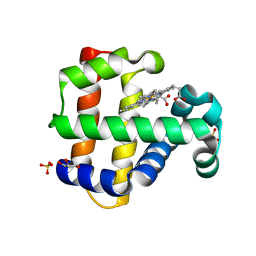 | | Crystal structure of dehaloperoxidase B in complex with 2,6-dibromophenol | | Descriptor: | 2,6-bis(bromanyl)phenol, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T. | | Deposit date: | 2021-03-13 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.554 Å) | | Cite: | Bridging the functional gap between reactivity and inhibition in dehaloperoxidase B from Amphitrite ornata: Mechanistic and structural studies with 2,4- and 2,6-dihalophenols.
J.Inorg.Biochem., 236, 2022
|
|
7M1I
 
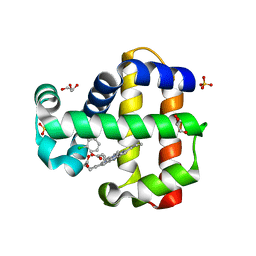 | | Crystal structure of dehaloperoxidase B in complex with 2,6-dichlorophenol | | Descriptor: | 2,6-dichlorophenol, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T. | | Deposit date: | 2021-03-13 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Bridging the functional gap between reactivity and inhibition in dehaloperoxidase B from Amphitrite ornata: Mechanistic and structural studies with 2,4- and 2,6-dihalophenols.
J.Inorg.Biochem., 236, 2022
|
|
