6L63
 
 | | Human Coagulation Factor XIIa (FXIIa) bound with the macrocyclic peptide F3 containing two (1S,2S)-2-ACHC residues | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL GROUP, ... | | Authors: | Sengoku, T, Katoh, T, Hirata, K, Suga, H, Ogata, K. | | Deposit date: | 2019-10-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ribosomal synthesis and de novo discovery of bioactive foldamer peptides containing cyclic beta-amino acids.
Nat.Chem., 12, 2020
|
|
6L6M
 
 | | HSP18.5 from E. histolytica | | Descriptor: | Heat shock protein hsp20 family putative | | Authors: | Kurre, D, Suguna, K. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.28000379 Å) | | Cite: | Network of Entamoeba histolytica HSP18.5 dimers formed by two overlapping [IV]-X-[IV] motifs.
Proteins, 2021
|
|
6LAZ
 
 | | the wildtype SAM-VI riboswitch bound to a N-mustard SAM analog M1 | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(2-hydroxyethyl)amino]-2-azaniumyl-butanoate, MAGNESIUM ION, RNA (55-MER), ... | | Authors: | Ren, A, Sun, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
6L8J
 
 | | Crystal structure of CYP97A3 mutant S290D/W300L/S304V in complex with retinal | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Protein LUTEIN DEFICIENT 5, chloroplastic, ... | | Authors: | Niu, G, Guo, Q, Liu, L. | | Deposit date: | 2019-11-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for plant lutein biosynthesis from alpha-carotene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1M4K
 
 | | Crystal structure of the human natural killer cell activator receptor KIR2DS2 (CD158j) | | Descriptor: | 1,2-ETHANEDIOL, KILLER CELL IMMUNOGLOBULIN-LIKE RECEPTOR 2DS2, SULFATE ION | | Authors: | Saulquin, X, Gastinel, L.N, Vivier, E. | | Deposit date: | 2002-07-03 | | Release date: | 2003-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the human natural killer cell
activating receptor KIR2DS2 (CD158j)
J.EXP.MED., 197, 2003
|
|
2F2X
 
 | | alpha-spectrin SH3 domain R21G mutant | | Descriptor: | SULFATE ION, Spectrin alpha chain, brain | | Authors: | Camara-Artigas, A, Conejero-Lara, F, Casares, S, Lopez-Mayorga, O, Vega, C. | | Deposit date: | 2005-11-18 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cooperative propagation of local stability changes from low-stability and high-stability regions in a SH3 domain
Proteins, 67, 2007
|
|
1M5K
 
 | |
2F3J
 
 | |
2F4G
 
 | | Triclinic cross-linked lysozyme soaked in bromoethanol 1M | | Descriptor: | 2-BROMOETHANOL, Lysozyme C, NITRATE ION | | Authors: | Prange, T, Salem, M. | | Deposit date: | 2005-11-23 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.654 Å) | | Cite: | On the edge of the denaturation process: Application of X-ray diffraction to barnase and lysozyme cross-linked crystals with denaturants in molar concentrations.
Biochim.Biophys.Acta, 1764, 2006
|
|
1M8B
 
 | | Solution structure of the C State of turkey ovomucoid at pH 2.5 | | Descriptor: | Ovomucoid | | Authors: | Song, J, Laskowski Jr, M, Qasim, M.A, Markley, J.L. | | Deposit date: | 2002-07-24 | | Release date: | 2002-09-04 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Two conformational states of Turkey ovomucoid third domain at low pH: three-dimensional structures, internal dynamics, and interconversion kinetics and thermodynamics.
Biochemistry, 42, 2003
|
|
1M8R
 
 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 7.4) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase A2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
1MNH
 
 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Davies, G.J, Wilkinson, A.J. | | Deposit date: | 1995-01-11 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
1M9U
 
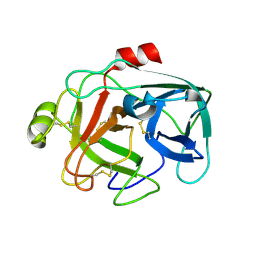 | |
2F7S
 
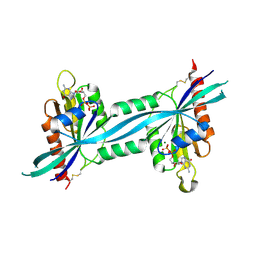 | | The crystal structure of human Rab27b bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-27B | | Authors: | Wang, J, Ismail, S, Shen, Y, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-01 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of human Rab27b
To be Published
|
|
2F81
 
 | | HIV-1 Protease mutant L90M complexed with inhibitor TMC114 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2005-12-01 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Effectiveness of Nonpeptide Clinical Inhibitor TMC-114 on HIV-1 Protease with Highly Drug Resistant Mutations D30N, I50V, and L90M.
J.Med.Chem., 49, 2006
|
|
1LV1
 
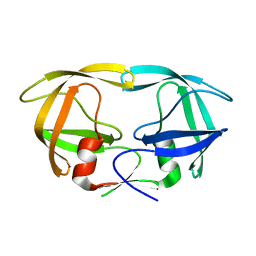 | | Crystal Structure Analysis of the non-active site mutant of tethered HIV-1 protease to 2.1A resolution | | Descriptor: | HIV-1 protease | | Authors: | Kumar, M, Kannan, K.K, Hosur, M.V, Bhavesh, N.S, Chatterjee, A, Mittal, R, Hosur, R.V. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effects of remote mutation on the autolysis of HIV-1 PR: X-ray and NMR investigations.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
2F7Z
 
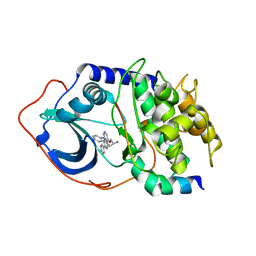 | | Protein Kinase A bound to (R)-1-(1H-Indol-3-ylmethyl)-2-(2-pyridin-4-yl-[1,7]naphtyridin-5-yloxy)-ehylamine | | Descriptor: | (1S)-1-(1H-INDOL-3-YLMETHYL)-2-(2-PYRIDIN-4-YL-[1,7]NAPHTYRIDIN-5-YLOXY)-EHYLAMINE, PKI, inhibitory peptide, ... | | Authors: | Li, Q, Woods, K.W, Thomas, S, Zhu, G.D, Packard, G, Fisher, J, Li, T, Gong, J, Dinges, J, Song, X, Abrams, J, Luo, Y, Johnson, E.F, Shi, Y, Liu, X, Klinghofer, V, Des Jong, R, Oltersdorf, T, Stoll, V.S, Jakob, C.G, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2005-12-01 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and structure-activity relationship of 3,4'-bispyridinylethylenes: discovery of a potent 3-isoquinolinylpyridine inhibitor of protein kinase B (PKB/Akt) for the treatment of cancer.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1MKU
 
 | |
2F9Q
 
 | | Crystal Structure of Human Cytochrome P450 2D6 | | Descriptor: | Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Rowland, P. | | Deposit date: | 2005-12-06 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Crystal Structure of Human Cytochrome P450 2D6
J.Biol.Chem., 281, 2006
|
|
2FA9
 
 | | The crystal structure of Sar1[H79G]-GDP provides insight into the coat-controlled GTP hydrolysis in the disassembly of COP II | | Descriptor: | GTP-binding protein SAR1b, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rao, Y, Huang, M, Yuan, C, Bian, C, Hou, X. | | Deposit date: | 2005-12-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Sar1[H79G]-GDP Which Provides Insight into the Coat-controlled GTP Hydrolysis in the Disassembly of COP II
Chin.J.Struct.Chem., 25, 2006
|
|
1LYA
 
 | | CRYSTAL STRUCTURES OF NATIVE AND INHIBITED FORMS OF HUMAN CATHEPSIN D: IMPLICATIONS FOR LYSOSOMAL TARGETING AND DRUG DESIGN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATHEPSIN D, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Baldwin, E.T, Bhat, T.N, Gulnik, S, Erickson, J.W. | | Deposit date: | 1993-04-22 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of native and inhibited forms of human cathepsin D: implications for lysosomal targeting and drug design.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1LYZ
 
 | |
1LZA
 
 | | DISSECTION OF PROTEIN-CARBOHYDRATE INTERACTIONS IN MUTANT HEN EGG-WHITE LYSOZYME COMPLEXES AND THEIR HYDROLYTIC ACTIVITY | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Maenaka, K, Matsushima, M, Song, H, Watanabe, K, Kumagai, I. | | Deposit date: | 1995-02-10 | | Release date: | 1995-05-08 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dissection of protein-carbohydrate interactions in mutant hen egg-white lysozyme complexes and their hydrolytic activity.
J.Mol.Biol., 247, 1995
|
|
2FDV
 
 | | Microsomal P450 2A6 with the inhibitor N-Methyl(5-(pyridin-3-yl)furan-2-yl)methanamine bound | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450 2A6, N-METHYL(5-(PYRIDIN-3-YL)FURAN-2-YL)METHANAMINE, ... | | Authors: | Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2005-12-14 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
1M30
 
 | | Solution structure of N-terminal SH3 domain from oncogene protein c-Crk | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-26 | | Release date: | 2003-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
