4LZG
 
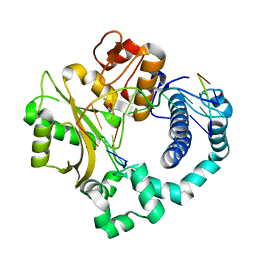 | | Binary complex of human DNA Polymerase Mu with DNA | | Descriptor: | CHLORIDE ION, DNA-directed DNA/RNA polymerase mu, GLYCEROL, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2013-07-31 | | Release date: | 2014-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Sustained active site rigidity during synthesis by human DNA polymerase mu.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1D93
 
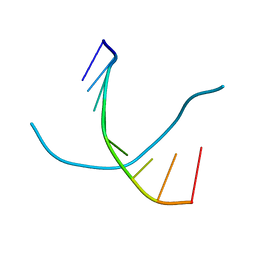 | |
4M04
 
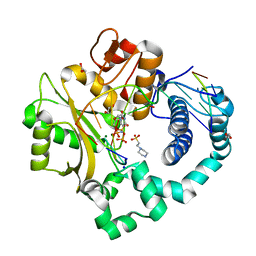 | | Human DNA Polymerase Mu ternary complex | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2013-08-01 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Sustained active site rigidity during synthesis by human DNA polymerase mu.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4MHG
 
 | | Crystal structure of ETV6 bound to a specific DNA sequence | | Descriptor: | Complementary Specific 14 bp DNA, Specific 14 bp DNA, Transcription factor ETV6 | | Authors: | Chan, A.C, De, S, Coyne III, H.J, Okon, M, Murphy, M.E, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2013-08-29 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Steric Mechanism of Auto-Inhibitory Regulation of Specific and Non-Specific DNA Binding by the ETS Transcriptional Repressor ETV6.
J.Mol.Biol., 426, 2014
|
|
3N78
 
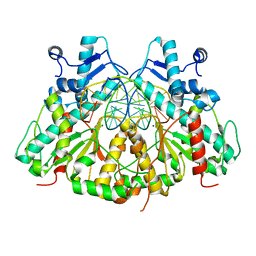 | | SgrAI bound to Secondary Site DNA and Mg(II) | | Descriptor: | DNA (5'-D(*AP*GP*TP*CP*CP*AP*CP*CP*GP*GP*GP*GP*GP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*TP*CP*CP*CP*CP*CP*GP*GP*TP*GP*GP*AP*CP*T)-3'), MAGNESIUM ION, ... | | Authors: | Horton, N.C, Little, E.J, Dunten, P.W. | | Deposit date: | 2010-05-26 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | New clues in the allosteric activation of DNA cleavage by SgrAI: structures of SgrAI bound to cleaved primary-site DNA and uncleaved secondary-site DNA.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1D02
 
 | | CRYSTAL STRUCTURE OF MUNI RESTRICTION ENDONUCLEASE IN COMPLEX WITH COGNATE DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*TP*TP*GP*GP*C)-3'), TYPE II RESTRICTION ENZYME MUNI | | Authors: | Deibert, M, Grazulis, S, Janulaitis, A, Siksnys, V, Huber, R. | | Deposit date: | 1999-09-08 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MunI restriction endonuclease in complex with cognate DNA at 1.7 A resolution.
EMBO J., 18, 1999
|
|
372D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
7S9N
 
 | | Binary complex of DNA Polymerase Beta with Fapy-dG in the template position | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*(FAP)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Freudenthal, B.D, Ryan, B.J, Smith, M.R. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural Dynamics of a Common Mutagenic Oxidative DNA Lesion in Duplex DNA and during DNA Replication.
J.Am.Chem.Soc., 144, 2022
|
|
7S9J
 
 | | Crystal Structure of DNA Polymerase Beta with Fapy-dG base-paired with a dC | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*(FAP)P*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*CP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Freudenthal, B.D, Ryan, B.J. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Dynamics of a Common Mutagenic Oxidative DNA Lesion in Duplex DNA and during DNA Replication.
J.Am.Chem.Soc., 144, 2022
|
|
7S9M
 
 | |
7S9K
 
 | | Crystal Structure of DNA Polymerase Beta with Fapy-dG base-paired with a dA | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*(FAP)P*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Freudenthal, B.D, Ryan, B.J. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Dynamics of a Common Mutagenic Oxidative DNA Lesion in Duplex DNA and during DNA Replication.
J.Am.Chem.Soc., 144, 2022
|
|
7S9L
 
 | |
7S9O
 
 | |
2DDG
 
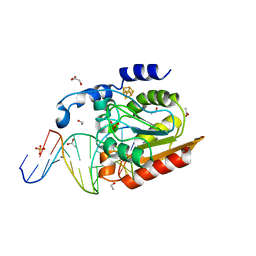 | | Crystal structure of uracil-DNA glycosylase in complex with AP:G containing DNA | | Descriptor: | 5'-D(*AP*TP*GP*TP*TP*GP*CP*(D1P)P*TP*TP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*AP*AP*GP*GP*CP*AP*AP*CP*A)-3', ACETATE ION, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Hoseki, J, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-28 | | Release date: | 2007-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
2DEM
 
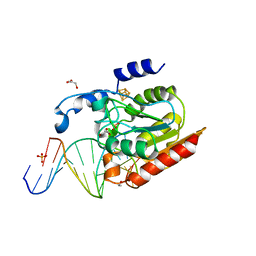 | | Crystal structure of Uracil-DNA glycosylase in complex with AP:A containing DNA | | Descriptor: | 5'-D(*AP*TP*GP*TP*TP*GP*CP*(D1P)P*TP*TP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*AP*AP*AP*GP*CP*AP*AP*CP*A)-3', DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Hoseki, J, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
6EHT
 
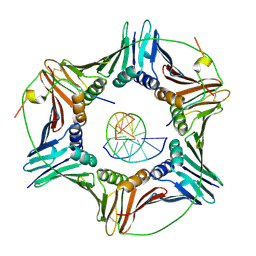 | | Modulation of PCNA sliding surface by p15PAF suggests a suppressive mechanism for cisplatin-induced DNA lesion bypass by pol eta holoenzyme | | Descriptor: | DNA (5'-D(P*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), PCNA-associated factor, ... | | Authors: | De March, M, Barrera-Vilarmau, S, Mentegari, E, Merino, N, Bressan, E, Maga, G, Crehuet, R, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | p15PAF binding to PCNA modulates the DNA sliding surface.
Nucleic Acids Res., 46, 2018
|
|
8Q3R
 
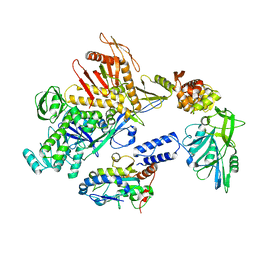 | | Cryo-EM structure of the DNA polymerase holoenzyme E9-A20-D4 of vaccinia virus | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component OPG148, Uracil-DNA glycosylase | | Authors: | Burmeister, W.P, Ballandras-Colas, A, Boettcher, B, Grimm, C. | | Deposit date: | 2023-08-04 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and flexibility of the DNA polymerase holoenzyme of vaccinia virus.
Plos Pathog., 20, 2024
|
|
5BNA
 
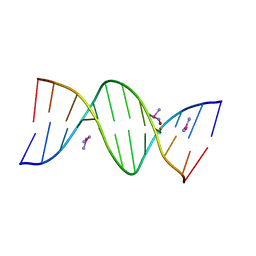 | | THE PRIMARY MODE OF BINDING OF CISPLATIN TO A B-DNA DODECAMER: C-G-C-G-A-A-T-T-C-G-C-G | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PLATINUM TRIAMINE ION | | Authors: | Wing, R.M, Pjura, P, Drew, H.R, Dickerson, R.E. | | Deposit date: | 1983-08-22 | | Release date: | 1983-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The primary mode of binding of cisplatin to a B-DNA dodecamer: C-G-C-G-A-A-T-T-C-G-C-G
EMBO J., 3, 1984
|
|
4LZD
 
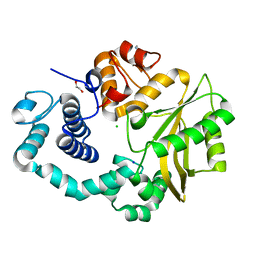 | | Human DNA polymerase mu- Apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-directed DNA/RNA polymerase mu, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2013-07-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Sustained active site rigidity during synthesis by human DNA polymerase mu.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2D3Y
 
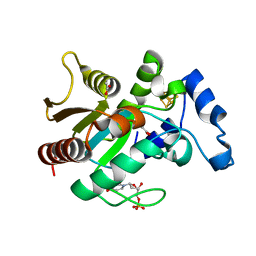 | | Crystal structure of uracil-DNA glycosylase from Thermus Thermophilus HB8 | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, ACETATE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
390D
 
 | | STRUCTURAL VARIABILITY AND NEW INTERMOLECULAR INTERACTIONS OF Z-DNA IN CRYSTALS OF D(PCPGPCPGPCPG) | | Descriptor: | DNA (5'-D(P*CP*GP*CP*GP*CP*G)-3') | | Authors: | Malinina, L, Tereshko, V, Ivanova, E, Subirana, J.A, Zarytova, V, Nekrasov, Y. | | Deposit date: | 1998-04-20 | | Release date: | 1998-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural variability and new intermolecular interactions of Z-DNA in crystals of d(pCpGpCpGpCpG).
Biophys.J., 74, 1998
|
|
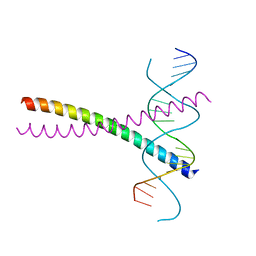
2DGC
 
 | |
1TUP
 
 | | TUMOR SUPPRESSOR P53 COMPLEXED WITH DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*AP*TP*TP*GP*GP*GP*CP*AP*AP*GP*TP*CP*TP*A P*GP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*CP*CP*TP*AP*GP*AP*CP*TP*TP*GP*CP*CP*CP*A P*AP*TP*TP*A)-3'), PROTEIN (P53 TUMOR SUPPRESSOR ), ... | | Authors: | Cho, Y, Gorina, S, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1995-07-11 | | Release date: | 1995-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations.
Science, 265, 1994
|
|
2P5O
 
 | |
1UAA
 
 | | E. COLI REP HELICASE/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), PROTEIN (ATP-DEPENDENT DNA HELICASE REP.) | | Authors: | Korolev, S, Waksman, G. | | Deposit date: | 1997-06-30 | | Release date: | 1998-07-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP.
Cell(Cambridge,Mass.), 90, 1997
|
|
