1TWM
 
 | | Interleukin-1 Beta Mutant F146Y | | Descriptor: | Interleukin-1 beta | | Authors: | Adamek, D.H, Capsar, D.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural and energetic consequences of mutations in a solvated hydrophobic cavity.
J.Mol.Biol., 346, 2005
|
|
1TXT
 
 | | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, ACETOACETYL-COENZYME A | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-07-06 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
1TOI
 
 | | Hydrocinnamic acid-bound structure of Hexamutant + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
3ZT4
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | 1,2-ETHANEDIOL, 5-[(E)-(2-OXO-2,3-DIHYDRO-1H-INDEN-1-YLIDENE)METHYL]-1,3-BENZODIOXOLE-4-CARBOXYLIC ACID, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design
Plos One, 7, 2012
|
|
3ZXE
 
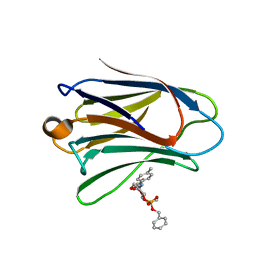 | | Crystal structure of Human Galectin-7 in complex with a galactose- benzylphosphate inhibitor | | Descriptor: | GALECTIN-7, METHYL 2-O-[(S)-(BENZYLOXY)(HYDROXY)PHOSPHORYL]-3-DEOXY-3-{[(4-METHYLPHENYL)CARBONYL]AMINO}-1-THIO-BETA-D-GALACTOPYRANOSIDE | | Authors: | Masuyer, G, Oberg, C.T, Leffler, H, Nilsson, U.J, Acharya, K.R. | | Deposit date: | 2011-08-10 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Inhibition Mechanism of Human Galectin-7 by a Novel Galactose-Benzylphosphate Inhibitor.
FEBS J., 279, 2012
|
|
3ZUI
 
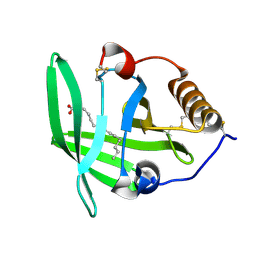 | | OMCI in complex with palmitoleic acid | | Descriptor: | COMPLEMENT INHIBITOR, PALMITOLEIC ACID | | Authors: | Roversi, P, Maillet, I, Togbe, D, Couillin, I, Quesniaux, V.F.J, Teixeira, M, Ahmat, N, Lissina, O, Boland, W, Ploss, K, Caesar, J.J.E, Leonhartsberger, S, Ryffel, B, Lea, S.M, Nunn, M.A. | | Deposit date: | 2011-07-19 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Bifunctional Lipocalin Ameliorates Murine Immune Complex-Induced Acute Lung Injury.
J.Biol.Chem., 288, 2013
|
|
3ZGH
 
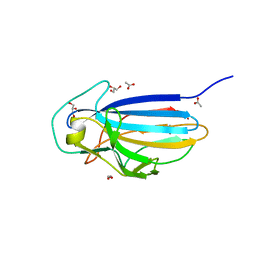 | | Crystal structure of the KRT10-binding region domain of the pneumococcal serine rich repeat protein PsrP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CELL WALL SURFACE ANCHOR FAMILY PROTEIN, ... | | Authors: | Schulte, T, Loefling, J, Mikaelsson, C, Kikhney, A, Hentrich, K, Diamante, A, Ebel, C, Normark, S, Svergun, D, Henriques-Normark, B, Achour, A. | | Deposit date: | 2012-12-17 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Basic Keratin 10-Binding Domain of the Virulence-Associated Pneumococcal Serine-Rich Protein Psrp Adopts a Novel Mscramm Fold.
Open Biol., 4, 2014
|
|
3Q9G
 
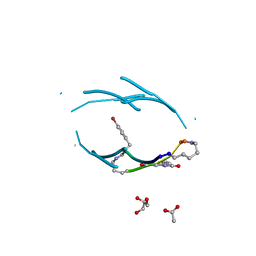 | | VQIVY segment from Alzheimer's tau displayed on 42-membered macrocycle scaffold | | Descriptor: | ACETIC ACID, Cyclic pseudo-peptide VQIV(4BF)(ORN)(HAO)KL(ORN), GLYCEROL | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
4BLI
 
 | | Galectin-3c in complex with Bisamido-thiogalactoside derivate 1 | | Descriptor: | (3-Deoxy-3-(3-methoxy-benzamido)-b-D-galactopyranosyl)-(3-deoxy-3-(3-methoxy-benzamido)-2-O-sulfo-b-D-galactopyranosyl)-sulfide, GALECTIN-3 | | Authors: | Noresson, A.L, Oberg, C.T, Engstrom, O, Hakansson, M, Logan, D.T, Leffler, H, Nilsson, U.J. | | Deposit date: | 2013-05-03 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Controlling Protein Conformation Through Electronic Fine-Tuning of Arginine-Arene Interactions: Synthetic, Structural, and Biological Studies
To be Published
|
|
1S9V
 
 | | Crystal structure of HLA-DQ2 complexed with deamidated gliadin peptide | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DQ(1) beta chain, ... | | Authors: | Kim, C.-Y, Quarsten, H, Bergseng, E, Khosla, C, Sollid, L.M. | | Deposit date: | 2004-02-05 | | Release date: | 2004-03-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4B0T
 
 | | Structure of the Pup Ligase PafA of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PUP--PROTEIN LIGASE | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.159 Å) | | Cite: | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
4BOM
 
 | | Structure of herpesvirus fusion glycoprotein B-bilayer complex revealing the protein-membrane and lateral protein-protein interaction | | Descriptor: | ENVELOPE GLYCOPROTEIN B | | Authors: | Maurer, U.E, Zeev-Ben-Mordehai, Z, Pandurangan, A.P, Cairns, T.M, Hannah, B.P, Whitbeck, J.C, Eisenberg, R.J, Cohen, G.H, Topf, M, Huiskonen, J.T, Grunewald, K. | | Deposit date: | 2013-05-21 | | Release date: | 2013-07-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (27 Å) | | Cite: | The structure of herpesvirus fusion glycoprotein B-bilayer complex reveals the protein-membrane and lateral protein-protein interaction.
Structure, 21, 2013
|
|
4CFR
 
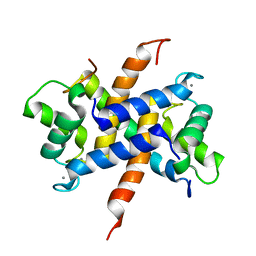 | | Ca-bound S100A4 C3S, C81S, C86S and F45W mutant complexed with non- muscle myosin IIA | | Descriptor: | CALCIUM ION, MYOSIN-9, PROTEIN S100-A4 | | Authors: | Duelli, A, Kiss, B, Lundholm, I, Bodor, A, Radnai, L, Petoukhov, M, Svergun, D, Nyitray, L, Katona, G. | | Deposit date: | 2013-11-19 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The C-Terminal Random Coil Region Tunes the Ca2+-Binding Affinity of S100A4 Through Conformational Activation.
Plos One, 9, 2014
|
|
1SL4
 
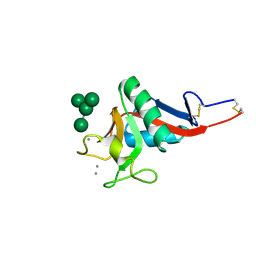 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with Man4 | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, mDC-SIGN1B type I isoform | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
4BV6
 
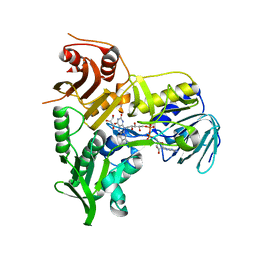 | | Refined crystal structure of the human Apoptosis inducing factor | | Descriptor: | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Hermoso, J.A, Ferreira, P, Villanueva, R, Medina, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Coenzyme Mediated Monomer-Dimer Transition of the Pro-Apoptotic Apoptosis Inducing Factor.
Biochemistry, 53, 2014
|
|
4BN6
 
 | |
1SQO
 
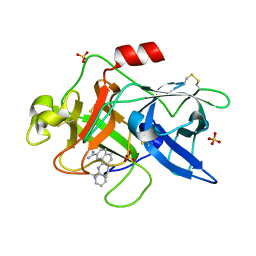 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 8-(PYRIMIDIN-2-YLAMINO)NAPHTHALENE-2-CARBOXIMIDAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhao, X, Mantei, R, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-19 | | Release date: | 2004-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1S2V
 
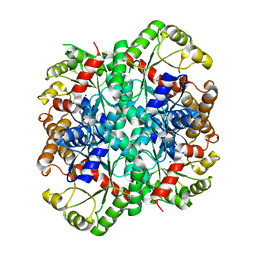 | | Crystal structure of phosphoenolpyruvate mutase complexed with Mg(II) | | Descriptor: | MAGNESIUM ION, Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1SIF
 
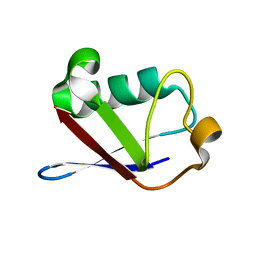 | | Crystal structure of a multiple hydrophobic core mutant of ubiquitin | | Descriptor: | ubiquitin | | Authors: | Benitez-Cardoza, C.G, Stott, K, Hirshberg, M, Went, H.M, Woolfson, D.N, Jackson, S.E. | | Deposit date: | 2004-02-29 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Exploring sequence/folding space: folding studies on multiple hydrophobic core mutants of ubiquitin
Biochemistry, 43, 2004
|
|
4C9T
 
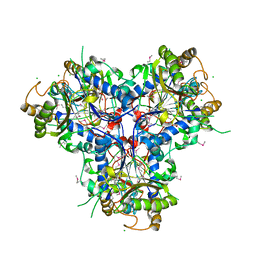 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 2.0 A RESOLUTION, SelenoMet derivative | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4CCS
 
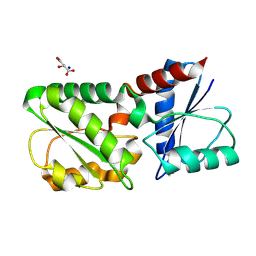 | | The structure of CbiX, the terminal Enzyme for Biosynthesis of Siroheme in Denitrifying Bacteria | | Descriptor: | 1,2-ETHANEDIOL, CBIX, D-MALATE, ... | | Authors: | Bali, S, Rollauer, S.E, Roversi, P, Raux-Deery, E, Lea, S.M, Warren, M.J, Ferguson, S.J. | | Deposit date: | 2013-10-25 | | Release date: | 2014-04-09 | | Last modified: | 2015-09-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Characterization of the 'Missing' Terminal Enzyme for Siroheme Biosynthesis in Alpha-Proteobacteria.
Mol.Microbiol., 92, 2014
|
|
3QER
 
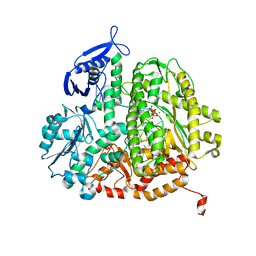 | | RB69 DNA Polymerase (L561A/S565G/Y567A) Ternary Complex with dATP Opposite Difluorotoluene Nucleoside | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-01-20 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for Differential Insertion Kinetics of dNMPs Opposite a Difluorotoluene Nucleotide Residue.
Biochemistry, 51, 2012
|
|
1UXE
 
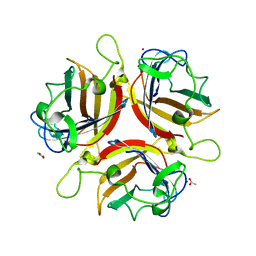 | | ADENOVIRUS AD37 FIBRE HEAD | | Descriptor: | ACETATE ION, FIBER PROTEIN, ZINC ION | | Authors: | Burmeister, W.P, Guilligay, D, Cusack, S, Wadell, G, Arnberg, N. | | Deposit date: | 2004-02-24 | | Release date: | 2004-07-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Species D Adenovirus Fiber Knobs and Their Sialic Acid Binding Sites
J.Virol., 78, 2004
|
|
3Q5L
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K 213 in the presence of 17-AEP-geldanamycin | | Descriptor: | (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-19-{[2-(pyrrolidin-1-yl)ethyl]amino}-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K 213 in the presence of 17-AEP-geldanamycin.
To be Published
|
|
3Q9I
 
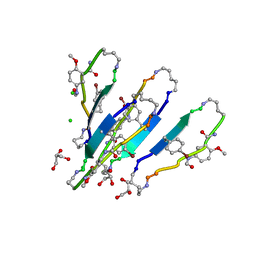 | | LVFFA segment from Alzheimer's Amyloid-Beta displayed on 42-membered macrocycle scaffold, bromide derivative | | Descriptor: | CHLORIDE ION, Cyclic pseudo-peptide LV(4BF)FA(ORN)(HAO)LK(ORN), GLYCEROL, ... | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
