6L5M
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
5IIA
 
 | | Crystal structure of red abalone egg VERL repeat 3 in complex with sperm lysin at 1.7 A resolution (crystal form I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Egg-lysin, Vitelline envelope sperm lysin receptor | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5INM
 
 | |
6S1R
 
 | |
5IBM
 
 | | Structure of S502P, a Cancer-Associated Mutation of the Oncogenic Phosphatase SHP2 | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Blacklow, S.C, Stams, T, Fodor, M, LaRochelle, J.R. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural and Functional Consequences of Three Cancer-Associated Mutations of the Oncogenic Phosphatase SHP2.
Biochemistry, 55, 2016
|
|
5IG3
 
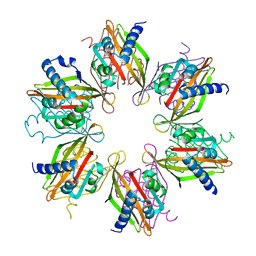 | | Crystal structure of the human CaMKII-alpha hub | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | McSpadden, E, Cao, Y.M, Bhattacharyya, M, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
6I4S
 
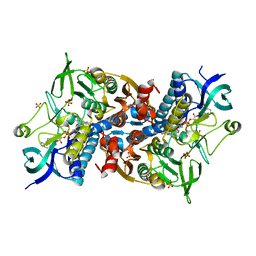 | | Crystal structure of the disease-causing R447G mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Szabo, E, Wilk, P, Hubert, A, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
6L5K
 
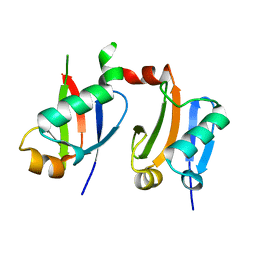 | | ARF5 Aux/IAA17 Complex | | Descriptor: | Auxin response factor 5, Auxin-responsive protein IAA17 | | Authors: | Ryu, K.S, Suh, J.Y, Cha, S.Y, Kim, Y.I, Park, C.K. | | Deposit date: | 2019-10-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Determinants of PB1 Domain Interactions in Auxin Response Factor ARF5 and Repressor IAA17.
J.Mol.Biol., 432, 2020
|
|
1BAB
 
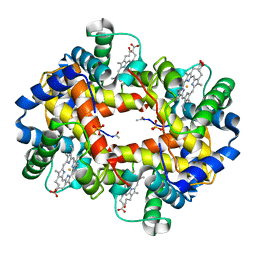 | |
5ILQ
 
 | | Crystal structure of truncated unliganded Aspartate Transcarbamoylase from Plasmodium falciparum | | Descriptor: | Aspartate carbamoyltransferase, GLYCEROL, SULFATE ION | | Authors: | Lunev, S, Bosch, S.S, Batista, F.D.A, Wrenger, C, Groves, M.R. | | Deposit date: | 2016-03-04 | | Release date: | 2016-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of truncated aspartate transcarbamoylase from Plasmodium falciparum.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6IU3
 
 | | Crystal structure of iron transporter VIT1 with zinc ions | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Taniguchi, R, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
1TCD
 
 | | TRYPANOSOMA CRUZI TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Maldonado, E, Soriano-Garcia, M, Cabrera, N, Garza-Ramos, G, Tuena De Gomez-Puyou, M, Gomez-Puyou, A, Perez-Montfort, R. | | Deposit date: | 1998-01-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Differences in the intersubunit contacts in triosephosphate isomerase from two closely related pathogenic trypanosomes.
J.Mol.Biol., 283, 1998
|
|
5IV1
 
 | | Solution Structure of DNA Dodecamer with 8-oxoguanine at 4th Position | | Descriptor: | DNA (5'-D(*CP*GP*CP*(8OG)P*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Miears, H.L, Gruber, D.R, Hoppins, J.J, Kiryutin, A.S, Kasymov, R.D, Yurkovskaya, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 8-Oxoguanine Affects DNA Backbone Conformation in the EcoRI Recognition Site and Inhibits Its Cleavage by the Enzyme.
Plos One, 11, 2016
|
|
6RZB
 
 | | Cryo-EM structure of mouse cytoplasmic dynein-1 microtubule binding domain bound to microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lacey, S.E, He, S, Scheres, S.H.W, Carter, A.P. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM of dynein microtubule-binding domains shows how an axonemal dynein distorts the microtubule.
Elife, 8, 2019
|
|
5IZ8
 
 | | Protein-protein interaction | | Descriptor: | ACE-ALA-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein, TRIETHYLENE GLYCOL | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5EUV
 
 | | Crystal structure of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d strain | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-D-galactosidase, ... | | Authors: | Rutkiewicz-Krotewicz, M, Bujacz, A, Pietrzyk, A.J, Sekula, B, Bujacz, G. | | Deposit date: | 2015-11-19 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8Q1K
 
 | | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Roske, Y, Daumke, O, Damme, M. | | Deposit date: | 2023-07-31 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation.
Nucleic Acids Res., 52, 2024
|
|
6IM3
 
 | | Crystal structure of a highly thermostable carbonic anhydrase from Persephonella marina EX-H1 | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CALCIUM ION, Carbonic anhydrase (Carbonate dehydratase), ... | | Authors: | Jin, M.S, Kim, S, Sung, J, Yeon, J, Choi, S.H. | | Deposit date: | 2018-10-22 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Highly Thermostable alpha-Carbonic Anhydrase from Persephonella marina EX-H1.
Mol.Cells, 42, 2019
|
|
5IZJ
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1411 | | Descriptor: | 4-(piperazin-1-yl)-7H-pyrrolo[2,3-d]pyrimidine, 47P-AZ1-DAR-DAR, 47P-AZ1-DAR-DAR-DAR, ... | | Authors: | Pflug, A, Enkvist, E, Uri, A, Engh, R.A. | | Deposit date: | 2016-03-25 | | Release date: | 2016-07-20 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bifunctional Ligands for Inhibition of Tight-Binding Protein-Protein Interactions.
Bioconjug.Chem., 27, 2016
|
|
8Q1X
 
 | | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Roske, Y, Daumke, O, Damme, M. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation.
Nucleic Acids Res., 52, 2024
|
|
5IZF
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1408 | | Descriptor: | 6J9-ZEU-DAR-ACA-DAR-NH2, SULFATE ION, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Enkvist, E, Uri, A, Engh, R.A. | | Deposit date: | 2016-03-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bifunctional Ligands for Inhibition of Tight-Binding Protein-Protein Interactions.
Bioconjug.Chem., 27, 2016
|
|
6RTL
 
 | | BACTERIOPHAGE SPP1 PROCAPSID-II PROTEIN | | Descriptor: | Major capsid protein | | Authors: | Ignatiou, A, Brasiles, S, El Sadek, M, Buerger, J, Mielke, T, Topf, M, Tavares, P. | | Deposit date: | 2019-05-24 | | Release date: | 2019-10-23 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural transitions during the scaffolding-driven assembly of a viral capsid.
Nat Commun, 10, 2019
|
|
6RZO
 
 | |
6IU6
 
 | | Crystal structure of cytoplasmic metal binding domain with nickel ions | | Descriptor: | NICKEL (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
5FX0
 
 | |
