7W2J
 
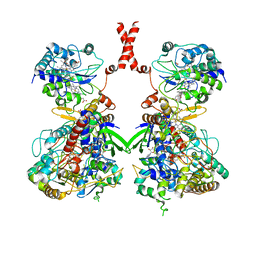 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
4JQP
 
 | |
4JE6
 
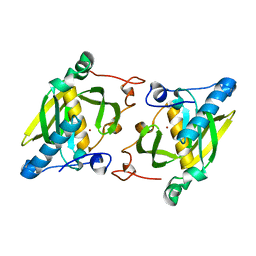 | | Crystal structure of a human-like mitochondrial peptide deformylase | | Descriptor: | Peptide deformylase 1A, chloroplastic/mitochondrial, ZINC ION | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8VCA
 
 | | Crystal Structure of plant Carboxylesterase 15 | | Descriptor: | Carboxylesterase 15, GLYCEROL | | Authors: | Palayam, M, Shabek, N. | | Deposit date: | 2023-12-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into strigolactone catabolism by carboxylesterases reveal a conserved conformational regulation.
Nat Commun, 15, 2024
|
|
6VS4
 
 | |
8VT8
 
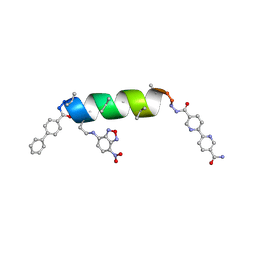 | | UIC-13-BIF-A4Dab NBD-Cl binding | | Descriptor: | 4-chloro-7-nitrobenzofurazan, ACETONITRILE, UIC-13-BIF-A4Dab-NBD-Cl | | Authors: | Ganatra, P. | | Deposit date: | 2024-01-26 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Diverse Proteomimetic Frameworks via Rational Design of pi-Stacking Peptide Tectons.
J.Am.Chem.Soc., 146, 2024
|
|
6GLW
 
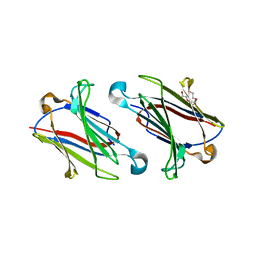 | | Structure of galectin-10 in complex with the Fab fragment of a Charcot-Leyden crystal solubilizing antibody, 1D11 | | Descriptor: | ACETATE ION, Fab 1D11 - Heavy chain, Fab 1D11 - Light chain, ... | | Authors: | Verstraete, K, Verschueren, K, Savvides, S.N. | | Deposit date: | 2018-05-23 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein crystallization promotes type 2 immunity and is reversible by antibody treatment.
Science, 364, 2019
|
|
6K16
 
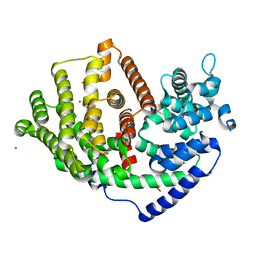 | |
8VPC
 
 | | UIC-13-BIF extension of UIC-1 | | Descriptor: | UIC-13-BIF | | Authors: | Ganatra, P. | | Deposit date: | 2024-01-16 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Diverse Proteomimetic Frameworks via Rational Design of pi-Stacking Peptide Tectons.
J.Am.Chem.Soc., 146, 2024
|
|
1UN2
 
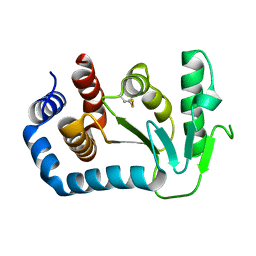 | | Crystal structure of circularly permuted CPDSBA_Q100T99: Preserved Global Fold and Local Structural Adjustments | | Descriptor: | THIOL-DISULFIDE INTERCHANGE PROTEIN | | Authors: | Manjasetty, B.A, Hennecke, J, Glockshuber, R, Heinemann, U. | | Deposit date: | 2003-09-03 | | Release date: | 2003-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Circularly Permuted Dsba(Q100T99): Preserved Global Fold and Local Structural Adjustments
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7A9U
 
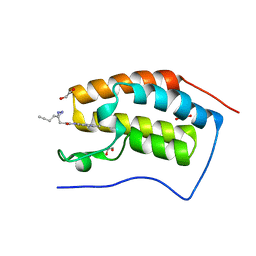 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 3-(3-(but-3-yn-1-yl)-3H-diazirin-3-yl)-N-(3-methyl-[1,2,4]triazolo[4,3-a]pyridin-8-yl)propanamide | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-(but-3-yn-1-yl)-3H-diazirin-3-yl)-N-(3-methyl-[1,2,4]triazolo[4,3-a]pyridin-8-yl)propanamide, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | A Photoaffinity-Based Fragment-Screening Platform for Efficient Identification of Protein Ligands.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8VBL
 
 | | Structure of bovine anti-HIV Fab ElsE6 | | Descriptor: | Bovine Fab ElsE6 heavy chain, Bovine Fab ElsE6 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2023-12-12 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire.
Plos Pathog., 20, 2024
|
|
3TAM
 
 | |
6K4G
 
 | | Crystal structure of the PI5P4Kbeta-GMPPNP complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Senda, M, Senda, T. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 2022
|
|
8VBR
 
 | | Structure of bovine anti-HIV Fab ElsE11 | | Descriptor: | Bovine Fab ElsE11 light chain, Bovine Fab Else11 heavy chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2023-12-12 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire.
Plos Pathog., 20, 2024
|
|
5V30
 
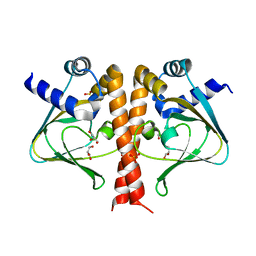 | | Crystal structure of the sensor domain of the transcriptional regulator HcpR from Porphyromonas Gingivalis | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator | | Authors: | Musayev, F.N, Belvin, B.R, Escalante, C.R, Turner, J, Lewis, J.P. | | Deposit date: | 2017-03-06 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Nitrosative stress sensing in Porphyromonas gingivalis: structure of and heme binding by the transcriptional regulator HcpR.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8QOB
 
 | |
6WAN
 
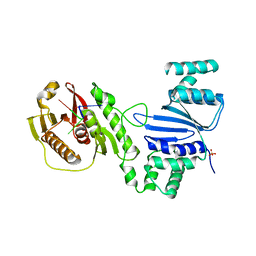 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor with the cyclic trinucleotide 3'3'3'-cAAA | | Descriptor: | Cyclic RNA (R(P*AP*AP*A), SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
4DZS
 
 | |
3TCZ
 
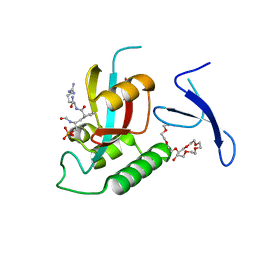 | | Human Pin1 bound to cis peptidomimetic inhibitor | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, N~2~-({(1R,2Z)-2-[(2R)-2-(formylamino)-3-(phosphonooxy)propylidene]cyclopentyl}carbonyl)-L-argininamide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Zhang, M, Zhang, Y. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Kinetic Analysis of Prolyl-isomerization/Phosphorylation Cross-Talk in the CTD Code.
Acs Chem.Biol., 7, 2012
|
|
3TGE
 
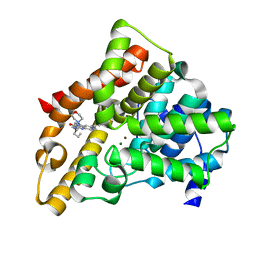 | | A novel series of potent and selective PDE5 inhibitor1 | | Descriptor: | 7-(6-methoxypyridin-3-yl)-3-{[2-(morpholin-4-yl)ethyl]amino}-1-(2-propoxyethyl)pyrido[3,4-b]pyrazin-2(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Han, S. | | Deposit date: | 2011-08-17 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Investigation of the pyrazinones as PDE5 inhibitors: Evaluation of regioisomeric projections into the solvent region.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
8GOD
 
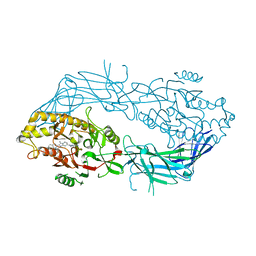 | | Co-crystal structure of Human Protein-arginine deiminase type-4 (PAD4) with small molecule inhibitor JBI-589 | | Descriptor: | Protein-arginine deiminase type-4, [(3~{R})-3-azanylpiperidin-1-yl]-[2-[1-[(4-fluorophenyl)methyl]indol-2-yl]-3-methyl-imidazo[1,2-a]pyridin-7-yl]methanone | | Authors: | Swaminathan, S, Birudukota, S, Vaithilingam, K, Kandan, S, Asaithambi, K, Kathiresan, N, Gosu, R, Rajagopal, S, Sadhu, N. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Alleviation of arthritis through prevention of neutrophil extracellular traps by an orally available inhibitor of protein arginine deiminase 4.
Sci Rep, 13, 2023
|
|
4TS9
 
 | |
8VBK
 
 | | Structure of bovine anti-HIV Fab ElsE5 | | Descriptor: | Bovine Fab ElsE5 heavy chain, Bovine Fab ElsE5 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2023-12-12 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire.
Plos Pathog., 20, 2024
|
|
6OAL
 
 | | Structure of human PARG complexed with JA2120 | | Descriptor: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-3,7-dihydro-1H-purine-2,6-dione, Poly(ADP-ribose) glycohydrolase | | Authors: | Brosey, C.A, Ahmed, Z, Warden, S, Tainer, J.A. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
