8YEW
 
 | | TRIP4 ASCH domain in unliganded form | | Descriptor: | Activating signal cointegrator 1 | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
9GDK
 
 | | Jumonji domain-containing protein 1C with crystallization epitope mutations L2440Y:G2444H | | Descriptor: | Probable JmjC domain-containing histone demethylation protein 2C | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-08-05 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To Be Published
|
|
8YFI
 
 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-TGAGGTACCTCA-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*A)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YFJ
 
 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-ATTGGATCCAAT-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*AP*TP*TP*GP*GP*AP*TP*CP*CP*AP*AP*T)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
7O6X
 
 | | Tankyrase 2 in complex with an inhibitor (OM-153) | | Descriptor: | N-[3-[5-(5-ethoxypyridin-2-yl)-4-(2-fluorophenyl)-1,2,4-triazol-3-yl]cyclobutyl]quinoxaline-5-carboxamide, Poly [ADP-ribose] polymerase tankyrase-2, ZINC ION | | Authors: | Sowa, S.T, Lehtio, L. | | Deposit date: | 2021-04-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of a 1,2,4-Triazole-Based Lead Tankyrase Inhibitor: Part II.
J.Med.Chem., 64, 2021
|
|
7NQO
 
 | | Mycobacterium tuberculosis Cytochrome P450 CYP121 in complex with lead compound 21 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[4-[2-(5-bromanyl-1~{H}-indol-3-yl)ethyl]pyrimidin-2-yl]morpholine, CHLORIDE ION, ... | | Authors: | Selvam, I.R. | | Deposit date: | 2021-03-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new strategy for hit generation: Novel in cellulo active inhibitors of CYP121A1 from Mycobacterium tuberculosis via a combined X-ray crystallographic and phenotypic screening approach (XP screen).
Eur.J.Med.Chem., 230, 2022
|
|
7NQM
 
 | | Mycobacterium tuberculosis Cytochrome P450 CYP121 in complex with lead compound 10 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(2-pyrimidin-4-ylethyl)-1~{H}-indole, CHLORIDE ION, ... | | Authors: | Selvam, I.R. | | Deposit date: | 2021-03-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new strategy for hit generation: Novel in cellulo active inhibitors of CYP121A1 from Mycobacterium tuberculosis via a combined X-ray crystallographic and phenotypic screening approach (XP screen).
Eur.J.Med.Chem., 230, 2022
|
|
7NQN
 
 | | Mycobacterium tuberculosis Cytochrome P450 CYP121 in complex with lead compound 14 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[4-[2-(1~{H}-indol-3-yl)ethyl]pyrimidin-2-yl]morpholine, CHLORIDE ION, ... | | Authors: | Selvam, I.R. | | Deposit date: | 2021-03-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new strategy for hit generation: Novel in cellulo active inhibitors of CYP121A1 from Mycobacterium tuberculosis via a combined X-ray crystallographic and phenotypic screening approach (XP screen).
Eur.J.Med.Chem., 230, 2022
|
|
7O3N
 
 | | Crystal Structure of AcrB Single Mutant - 2 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter | | Authors: | Ababou, A. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.561 Å) | | Cite: | Crystal Structure of AcrB Single Mutant - 2
To Be Published
|
|
3EQM
 
 | |
7O1F
 
 | |
7O1E
 
 | | Crystal structure of PCNA from Chaetomium thermophilum | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Alphey, M.A, MacNeill, S, Yang, D. | | Deposit date: | 2021-03-29 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Non-canonical binding of the Chaetomium thermophilum PolD4 N-terminal PIP motif to PCNA involves Q-pocket and compact 2-fork plug interactions but no 3 10 helix.
Febs J., 290, 2023
|
|
7O3M
 
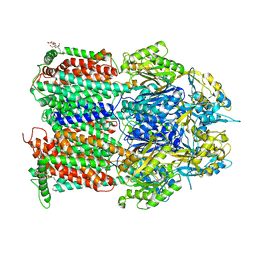 | | Crystal Structure of AcrB Single Mutant - 1 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter | | Authors: | Ababou, A. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.551 Å) | | Cite: | Crystal Structure of AcrB Single Mutant - 1
To Be Published
|
|
7O3L
 
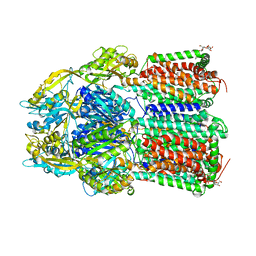 | | Crystal Structure of AcrB Double Mutant | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter | | Authors: | Ababou, A. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.526 Å) | | Cite: | Crystal Structure of AcrB Double Mutant
To Be Published
|
|
3EYU
 
 | |
3FCC
 
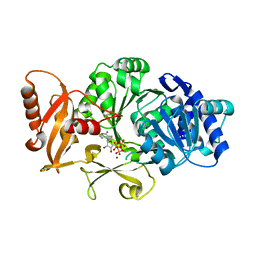 | | CRYSTAL STRUCTURE OF DLTA PROTEIN IN COMPLEX WITH ATP and MAGNESIUM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-alanine--poly(phosphoribitol) ligase subunit 1, MAGNESIUM ION | | Authors: | Osman, K.T, Du, L, He, Y, Luo, Y. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of Bacillus cereus D-alanyl carrier protein ligase (DltA) in complex with ATP.
J.Mol.Biol., 388, 2009
|
|
1OSV
 
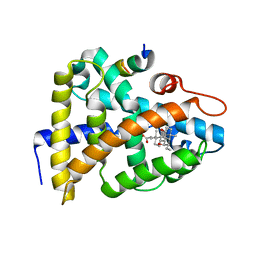 | | STRUCTURAL BASIS FOR BILE ACID BINDING AND ACTIVATION OF THE NUCLEAR RECEPTOR FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
5AGQ
 
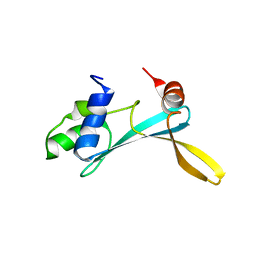 | | Solution structure of the TAM domain of human TIP5 BAZ2A involved in epigenetic regulation of rRNA genes | | Descriptor: | BROMODOMAIN ADJACENT TO ZINC FINGER DOMAIN PROTEIN 2A | | Authors: | Anosova, I, Melnik, S, Tripsianes, K, Kateb, F, Grummt, I, Sattler, M. | | Deposit date: | 2015-02-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Novel RNA Binding Surface of the Tam Domain of Tip5/Baz2A Mediates Epigenetic Regulation of Rrna Genes.
Nucleic Acids Res., 43, 2015
|
|
5C5S
 
 | |
6GC3
 
 | |
7T49
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 10c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7SSE
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with CYCA-117-70 | | Descriptor: | DDB1- and CUL4-associated factor 1, N-[(3R)-1-(3-fluorophenyl)piperidin-3-yl]-6-(morpholin-4-yl)pyrimidin-4-amine | | Authors: | Kimani, S, Owen, J, Li, A, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Shahani, V.M, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of a Novel DCAF1 Ligand Using a Drug-Target Interaction Prediction Model: Generalizing Machine Learning to New Drug Targets.
J.Chem.Inf.Model., 63, 2023
|
|
7T40
 
 | | Structure of MERS 3CL protease in complex with inhibitor 10c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T4B
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 14c | | Descriptor: | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
2D89
 
 | | Solution structure of the CH domain from human EH domain binding protein 1 | | Descriptor: | EHBP1 protein | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-02 | | Release date: | 2006-06-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain from human EH domain binding protein 1
To be Published
|
|
