7BFL
 
 | | X-ray structure of SS-RNase-2 des116-120 | | Descriptor: | Angiogenin-1 | | Authors: | Sica, F, Russo Krauss, I, Troisi, R. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | The structural features of an ancient ribonuclease from Salmo salar reveal an intriguing case of auto-inhibition.
Int.J.Biol.Macromol., 182, 2021
|
|
7BFK
 
 | | X-ray structure of SS-RNase-2 | | Descriptor: | Angiogenin-1 | | Authors: | Sica, F, Russo Krauss, I, Troisi, R. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The structural features of an ancient ribonuclease from Salmo salar reveal an intriguing case of auto-inhibition.
Int.J.Biol.Macromol., 182, 2021
|
|
1ZH5
 
 | | Structural basis for recognition of UUUOH 3'-terminii of nascent RNA pol III transcripts by La autoantigen | | Descriptor: | 5'-R(*UP*GP*CP*UP*GP*UP*UP*UP*U)-3', Lupus La protein, SULFATE ION | | Authors: | Teplova, M, Yuan, Y.R, Ilin, S, Malinina, L, Phan, A.T, Teplov, A, Patel, D.J. | | Deposit date: | 2005-04-22 | | Release date: | 2006-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Recognition and Sequestration of UUU(OH) 3' Temini of Nascent RNA Polymerase III Transcripts by La, a Rheumatic Disease Autoantigen.
Mol.Cell, 21, 2006
|
|
5KPY
 
 | | Structure of a 5-hydroxytryptophan aptamer | | Descriptor: | 5-hydroxy-L-tryptophan, 5-hydroxytryptophan RNA aptamer, IRIDIUM HEXAMMINE ION, ... | | Authors: | Batey, R.T, Porter, E, Merck, M. | | Deposit date: | 2016-07-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recurrent RNA motifs as scaffolds for genetically encodable small-molecule biosensors.
Nat. Chem. Biol., 13, 2017
|
|
3C3Z
 
 | | Crystal structure of HIV-1 subtype F DIS extended duplex RNA bound to ribostamycin | | Descriptor: | HIV-1 subtype F genomic RNA, RIBOSTAMYCIN | | Authors: | Freisz, S, Lang, K, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-01-29 | | Release date: | 2008-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
4JRD
 
 | | Crystal structure of the parallel double-stranded helix of poly(A) RNA | | Descriptor: | AMMONIUM ION, RNA (5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Safaee, N, Noronha, A.M, Kozlov, G, Rodionov, D, Wilds, C.J, Sheldrick, G.M, Gehring, K. | | Deposit date: | 2013-03-21 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the parallel duplex of poly(A) RNA: evaluation of a 50 year-old prediction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2JTP
 
 | |
5DOT
 
 | | Crystal Structure of Human Carbamoyl phosphate synthetase I (CPS1), apo form | | Descriptor: | 1,2-ETHANEDIOL, Carbamoyl-phosphate synthase [ammonia], mitochondrial, ... | | Authors: | Polo, L.M, de Cima, S, Fita, I, Rubio, V. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis.
Sci Rep, 5, 2015
|
|
1ARO
 
 | | T7 RNA POLYMERASE COMPLEXED WITH T7 LYSOZYME | | Descriptor: | MERCURY (II) ION, T7 LYSOZYME, T7 RNA POLYMERASE | | Authors: | Steitz, T, Jeruzalmi, D. | | Deposit date: | 1997-08-08 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of T7 RNA polymerase complexed to the transcriptional inhibitor T7 lysozyme.
EMBO J., 17, 1998
|
|
2E1B
 
 | |
4O2D
 
 | |
1WW1
 
 | |
7KUM
 
 | |
7KUP
 
 | |
8D5K
 
 | |
1C8M
 
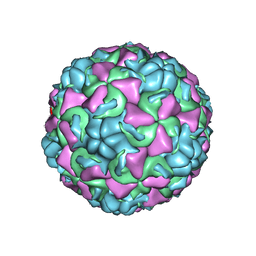 | | REFINED CRYSTAL STRUCTURE OF HUMAN RHINOVIRUS 16 COMPLEXED WITH VP63843 (PLECONARIL), AN ANTI-PICORNAVIRAL DRUG CURRENTLY IN CLINICAL TRIALS | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, HUMAN RHINOVIRUS 16 COAT PROTEIN, ZINC ION | | Authors: | Chakravarty, S, Bator, C.M, Pevear, D.C, Diana, G.D, Rossmann, M.G. | | Deposit date: | 2000-05-26 | | Release date: | 2000-11-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | THE REFINED STRUCTURE OF A PICORNAVIRUS INHIBITOR CURRENTLY IN CLINICAL TRIALS, WHEN COMPLEXED WITH HUMAN RHINOVIRUS 16
to be published
|
|
2GHQ
 
 | |
2GHT
 
 | |
5DOU
 
 | | Crystal Structure of Human Carbamoyl phosphate synthetase I (CPS1), ligand-bound form | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | de Cima, S, Polo, L.M, Fita, I, Rubio, V. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis.
Sci Rep, 5, 2015
|
|
6AGG
 
 | |
4LJ0
 
 | | Nab2 Zn fingers complexed with polyadenosine | | Descriptor: | ACETATE ION, MAGNESIUM ION, Nab2, ... | | Authors: | Stewart, M, Kuhlmann, S.I, Valkov, E. | | Deposit date: | 2013-07-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the molecular recognition of polyadenosine RNA by Nab2 Zn fingers.
Nucleic Acids Res., 42, 2014
|
|
6VTE
 
 | | Naegleria gruberi RNA Ligase K170M mutant with AMP and Mn | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
1Z3C
 
 | | Encephalitozooan cuniculi mRNA Cap (Guanine-N7) Methyltransferasein complexed with AzoAdoMet | | Descriptor: | S-5'-AZAMETHIONINE-5'-DEOXYADENOSINE, mRNA CAPPING ENZYME | | Authors: | Hausmann, S, Zhang, S, Fabrega, C, Schneller, S.W, Lima, C.D, Shuman, S. | | Deposit date: | 2005-03-11 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Encephalitozoon cuniculi mRNA cap (guanine N-7) methyltransferase: methyl acceptor specificity, inhibition BY S-adenosylmethionine analogs, and structure-guided mutational analysis.
J.Biol.Chem., 280, 2005
|
|
6VT6
 
 | | Naegleria gruberi RNA ligase K170A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT9
 
 | | Naegleria gruberi RNA ligase E227A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
