3UVE
 
 | |
3CSE
 
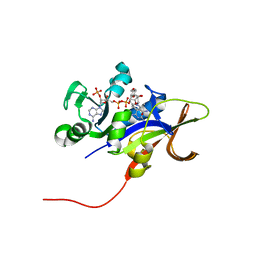 | | Candida glabrata Dihydrofolate Reductase complexed with NADPH and 2,4-diamino-5-(3-(2,5-dimethoxyphenyl)prop-1-ynyl)-6-ethylpyrimidine (UCP120B) | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, J, Anderson, A.C. | | Deposit date: | 2008-04-09 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-guided development of efficacious antifungal agents targeting Candida glabrata dihydrofolate reductase.
Chem.Biol., 15, 2008
|
|
4TU5
 
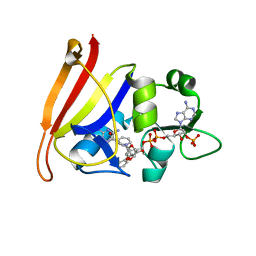 | |
2G5W
 
 | | X-ray crystal structure of Arabidopsis thaliana 12-oxophytodienoate reductase isoform 3 (AtOPR3) in complex with 8-iso prostaglandin A1 and its cofactor, flavin mononucleotide. | | Descriptor: | (8S,12S)-15S-HYDROXY-9-OXOPROSTA-10Z,13E-DIEN-1-OIC ACID, 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE | | Authors: | Han, B.W, Malone, T.E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Fox, B.G, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-23 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.576 Å) | | Cite: | Crystal structure of Arabidopsis thaliana 12-oxophytodienoate reductase isoform 3 in complex with 8-iso prostaglandin A(1).
Proteins, 79, 2011
|
|
6MUB
 
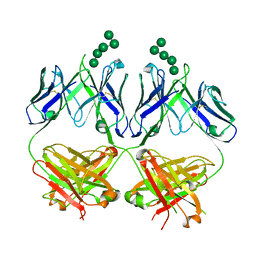 | | Anti-HIV-1 Fab 2G12 + Man5 re-refinement | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Wilson, I.A, Calarese, D.A, Stanfield, R.L. | | Deposit date: | 2018-10-22 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1GLQ
 
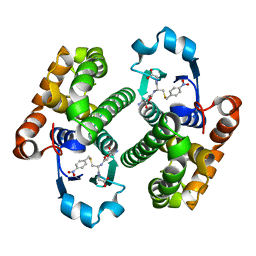 | |
1XVW
 
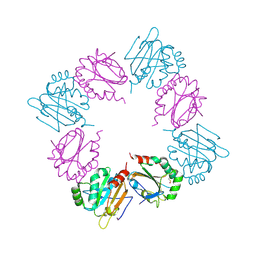 | | Crystal Structure of AhpE from Mycobacterium tuberculosis, a 1-Cys peroxiredoxin | | Descriptor: | Hypothetical protein Rv2238c/MT2298 | | Authors: | Li, S, Peterson, N.A, Kim, M.Y, Kim, C.Y, Hung, L.W, Yu, M, Lekin, T, Segelke, B.W, Lott, J.S, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-10-28 | | Release date: | 2005-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of AhpE from Mycobacterium tuberculosis, a 1-Cys Peroxiredoxin
J.Mol.Biol., 346, 2005
|
|
1QXS
 
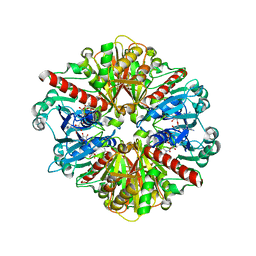 | | CRYSTAL STRUCTURE OF Trypanosoma cruzi GLYCERALDEHYDE-3- PHOSPHATE DEHYDROGENASE COMPLEXED WITH AN ANALOGUE OF 1,3- BisPHOSPHO-D-GLYCERIC ACID | | Descriptor: | 3-HYDROXY-2-OXO-4-PHOPHONOXY- BUTYL)-PHOSPHONIC ACID, Glyceraldehyde 3-phosphate dehydrogenase, glycosomal, ... | | Authors: | Castilho, M.S, Pavao, F, Oliva, G. | | Deposit date: | 2003-09-08 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of Trypanosoma cruzi glyceraldehyde-3-phosphate dehydrogenase complexed with an analogue of 1,3-bisphospho-d-glyceric acid.
Eur.J.Biochem., 270, 2003
|
|
6B1O
 
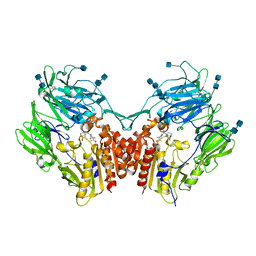 | | The structure of DPP4 in complex with Vildagliptin Analog | | Descriptor: | (2S)-2-amino-1-[(1S,3S,5S)-3-(aminomethyl)-2-azabicyclo[3.1.0]hexan-2-yl]-2-[(1r,3R,5S,7S)-3,5-dihydroxytricyclo[3.3.1.1~3,7~]decan-1-yl]ethan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2017-09-18 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A comparative study of the binding properties, dipeptidyl peptidase-4 (DPP-4) inhibitory activity and glucose-lowering efficacy of the DPP-4 inhibitors alogliptin, linagliptin, saxagliptin, sitagliptin and vildagliptin in mice.
Endocrinol Diabetes Metab, 1, 2018
|
|
3AK1
 
 | | Superoxide dismutase from Aeropyrum pernix K1, apo-form | | Descriptor: | 1,2-ETHANEDIOL, Superoxide dismutase [Mn/Fe] | | Authors: | Nakamura, T, Uegaki, K. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of the cambialistic superoxide dismutase from Aeropyrum pernix K1 - insights into the enzyme mechanism and stability
Febs J., 278, 2011
|
|
5F1A
 
 | | The Crystal Structure of Salicylate Bound to Human Cyclooxygenase-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-HYDROXYBENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lucido, M.J, Orlando, B.J, Malkowski, M.G. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of Aspirin-Acetylated Human Cyclooxygenase-2: Insight into the Formation of Products with Reversed Stereochemistry.
Biochemistry, 55, 2016
|
|
5SUX
 
 | |
1XXU
 
 | | Crystal Structure of AhpE from Mycrobacterium tuberculosis, a 1-Cys peroxiredoxin | | Descriptor: | Hypothetical protein Rv2238c/MT2298 | | Authors: | Li, S, Peterson, N.A, Kim, M.Y, Kim, C.Y, Hung, L.W, Yu, M, Lekin, T, Segelke, B.W, Lott, J.S, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-08 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of AhpE from Mycobacterium tuberculosis, a 1-Cys Peroxiredoxin
J.Mol.Biol., 346, 2005
|
|
2H7R
 
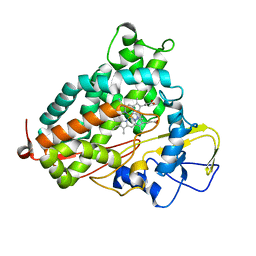 | |
1GLP
 
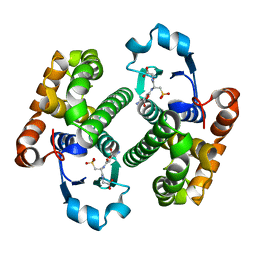 | |
3IS4
 
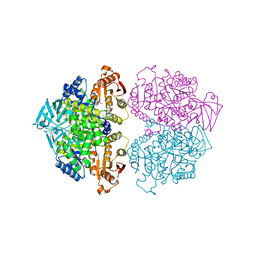 | | Crystal structure of Leishmania mexicana pyruvate kinase (LmPYK)in complex with 1,3,6,8-pyrenetetrasulfonic acid | | Descriptor: | GLYCEROL, Pyruvate kinase, pyrene-1,3,6,8-tetrasulfonic acid | | Authors: | Walkinshaw, M.D, Morgan, H.P. | | Deposit date: | 2009-08-25 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An improved strategy for the crystallization of Leishmania mexicana pyruvate kinase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3AK3
 
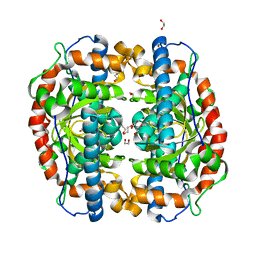 | | Superoxide dismutase from Aeropyrum pernix K1, Fe-bound form | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Superoxide dismutase [Mn/Fe] | | Authors: | Nakamura, T, Uegaki, K. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of the cambialistic superoxide dismutase from Aeropyrum pernix K1 - insights into the enzyme mechanism and stability
Febs J., 278, 2011
|
|
3BS6
 
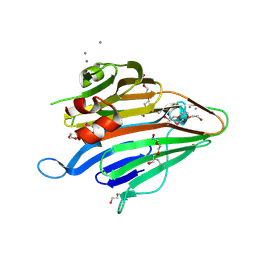 | |
1U18
 
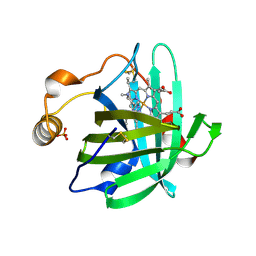 | |
3QY1
 
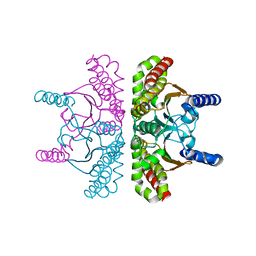 | | 1.54A Resolution Crystal Structure of a Beta-Carbonic Anhydrase from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 1.54A Resolution Crystal Structure of a Beta-Carbonic Anhydrase from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
To be Published
|
|
2JBV
 
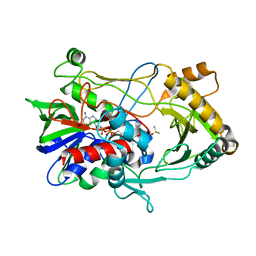 | | Crystal structure of choline oxidase reveals insights into the catalytic mechanism | | Descriptor: | CHOLINE OXIDASE, DIMETHYL SULFOXIDE, UNKNOWN ATOM OR ION, ... | | Authors: | Lountos, G.T, Fan, F, Gadda, G, Orville, A.M. | | Deposit date: | 2006-12-13 | | Release date: | 2007-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Role of Glu312 in Binding and Positioning of the Substrate for the Hydride Transfer Reaction in Choline Oxidase.
Biochemistry, 47, 2008
|
|
3AK2
 
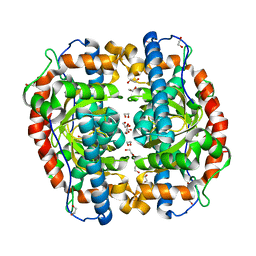 | | Superoxide dismutase from Aeropyrum pernix K1, Mn-bound form | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Superoxide dismutase [Mn/Fe] | | Authors: | Nakamura, T, Uegaki, K. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the cambialistic superoxide dismutase from Aeropyrum pernix K1 - insights into the enzyme mechanism and stability
Febs J., 278, 2011
|
|
1Z5Y
 
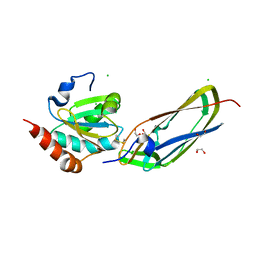 | | Crystal Structure Of The Disulfide-Linked Complex Between The N-Terminal Domain Of The Electron Transfer Catalyst DsbD and The Cytochrome c Biogenesis Protein CcmG | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Thiol:disulfide interchange protein dsbD, ... | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Gruetter, M.G, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-03-21 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Basis and Kinetics of DsbD-Dependent Cytochrome c Maturation
STRUCTURE, 13, 2005
|
|
3FPL
 
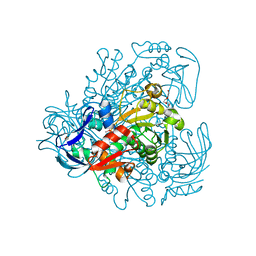 | | Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of C. beijerinckii ADH by T. brockii ADH | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Felix, F, Goihberg, E, Shimon, L, Burstein, Y. | | Deposit date: | 2009-01-05 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural properties of chimeras constructed by exchange of cofactor-binding domains in alcohol dehydrogenases from thermophilic and mesophilic microorganisms
Biochemistry, 49, 2010
|
|
1ZGK
 
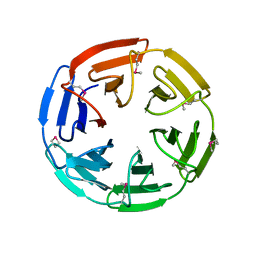 | | 1.35 angstrom structure of the Kelch domain of Keap1 | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Li, X, Bottoms, C.A, Hannink, M, Beamer, L.J. | | Deposit date: | 2005-04-21 | | Release date: | 2005-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conserved solvent and side-chain interactions in the 1.35 Angstrom structure of the Kelch domain of Keap1.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
