6S0W
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel and kanamycin B sulfate | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, DI(HYDROXYETHYL)ETHER, Kanamycin B dioxygenase, ... | | Authors: | Mrugala, B, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
4ZSU
 
 | |
7CEQ
 
 | |
7CER
 
 | | Crystal structure of D-cycloserine-bound form of cysteine desulfurase SufS H121A from Bacillus subtilis | | Descriptor: | Cysteine desulfurase SufS, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
6S0U
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
7CEP
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase SufS from Bacillus subtilis | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Yasuhiro, T, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
7CEO
 
 | |
7CES
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase SufS H121A from Bacillus subtilis | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
5KK3
 
 | | Atomic Resolution Structure of Monomorphic AB42 Amyloid Fibrils | | Descriptor: | Beta-amyloid protein 42 | | Authors: | Colvin, M.T, Silvers, R, Zhe Ni, Q, Can, T.V, Sergeyev, I, Rosay, M, Donovan, K.J, Michael, B, Wall, J, Linse, S, Griffin, R.G. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic Resolution Structure of Monomorphic A beta 42 Amyloid Fibrils.
J.Am.Chem.Soc., 138, 2016
|
|
3EEF
 
 | | Crystal structure of N-carbamoylsarcosine amidase from thermoplasma acidophilum | | Descriptor: | N-carbamoylsarcosine amidase related protein, ZINC ION | | Authors: | Luo, H.-B, Zheng, H, Chruszcz, M, Zimmerman, M.D, Skarina, T, Egorova, O, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and molecular modeling study of N-carbamoylsarcosine amidase Ta0454 from Thermoplasma acidophilum.
J.Struct.Biol., 169, 2010
|
|
6S0T
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel, sulfate, soaked with iodide | | Descriptor: | IODIDE ION, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Cymborowski, M.T, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
3EH9
 
 | | Crystal structure of death associated protein kinase complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Death-associated protein kinase 1, SULFATE ION | | Authors: | McNamara, L.K, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into nucleotide recognition by human death-associated protein kinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6SDF
 
 | | N-terminal SH3 domain of Grb2 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Growth factor receptor-bound protein 2 | | Authors: | Bolgov, A.A, Korban, S.A, Luzik, D.A, Rogacheva, O.N, Zhemkov, V.A, Kim, M, Skrynnikov, N.R, Bezprozvanny, I.B. | | Deposit date: | 2019-07-26 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the SH3 domain of growth factor receptor-bound protein 2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
3EHA
 
 | |
9EBZ
 
 | |
6RG2
 
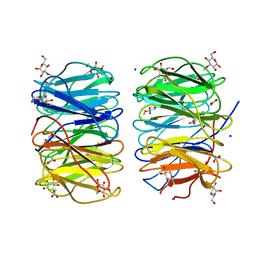 | | Photorhabdus laumondii lectin PLL2 in complex with 3-O-methyl-D-glucose | | Descriptor: | 3-O-methyl-alpha-D-glucopyranose, 3-O-methyl-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Houser, J, Fujdiarova, E, Wimmerova, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Heptabladed beta-propeller lectins PLL2 and PHL from Photorhabdus spp. recognize O-methylated sugars and influence the host immune system.
Febs J., 288, 2021
|
|
6RGG
 
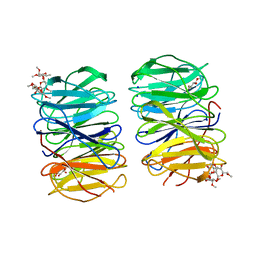 | | Photorhabdus laumondii lectin PLL2 in complex with O-methylated PGL-1-derived disaccharide | | Descriptor: | 1,2-ETHANEDIOL, 3,6-O-dimethyl-D-glucose, 6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose, ... | | Authors: | Houser, J, Fujdiarova, E, Wimmerova, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Heptabladed beta-propeller lectins PLL2 and PHL from Photorhabdus spp. recognize O-methylated sugars and influence the host immune system.
Febs J., 288, 2021
|
|
5LUD
 
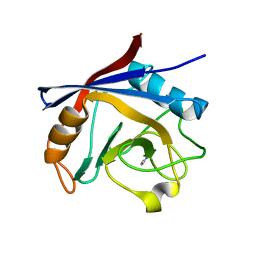 | | Structure of Cyclophilin A in complex with 2,3-Diaminopyridine | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, pyridine-2,3-diamine | | Authors: | McNae, I.W, Nowicki, M.W, Blackburn, E.A, Wear, M.A, Walkinshaw, M.D. | | Deposit date: | 2016-09-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Thermo-kinetic analysis space expansion for cyclophilin-ligand interactions - identification of a new nonpeptide inhibitor using BiacoreTM T200.
FEBS Open Bio, 7, 2017
|
|
6NFF
 
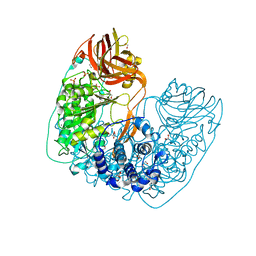 | | Structure of X-prolyl dipeptidyl aminopeptidase from Lactobacillus helveticus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Juers, D.H, Bratt, N.J, Ojennus, D.D. | | Deposit date: | 2018-12-20 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of a prolyl aminodipeptidase (PepX) from Lactobacillus helveticus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8EJN
 
 | | Structure of dehaloperoxidase A in complex with 2,4-dichlorophenol | | Descriptor: | 2,4-dichlorophenol, DIMETHYL SULFOXIDE, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Franzen, S. | | Deposit date: | 2022-09-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Comparative study of the binding and activation of 2,4-dichlorophenol by dehaloperoxidase A and B.
J.Inorg.Biochem., 247, 2023
|
|
6NOV
 
 | | A Fab derived from ixekizumab | | Descriptor: | DODECAETHYLENE GLYCOL, Fab Heavy Chain, Fab Light Chain | | Authors: | Durbin, J.D, Clawson, D.K, Lu, F, Tian, Y, Lu, J, Schmitt, M, Atwell, S. | | Deposit date: | 2019-01-16 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Development of tibulizumab, a tetravalent bispecific antibody targeting BAFF and IL-17A for the treatment of autoimmune disease.
Mabs, 11, 2019
|
|
5LRQ
 
 | | BRD4 in complex with ERK5 inhibitor XMD8-92 | | Descriptor: | 2-{[2-ethoxy-4-(4-hydroxypiperidin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Martin, M.P, Noble, M.E.M. | | Deposit date: | 2016-08-19 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a novel orally bioavailable ERK5 inhibitor with selectivity over p38 alpha and BRD4.
Eur.J.Med.Chem., 178, 2019
|
|
5LYE
 
 | |
9DIN
 
 | |
6VPB
 
 | |
