2O0H
 
 | | T4 gp17 ATPase domain mutant complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA packaging protein Gp17 | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2006-11-27 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Structure of the ATPase that Powers DNA Packaging into Bacteriophage T4 Procapsids
MOL.CELL, 25, 2007
|
|
2IQC
 
 | | Crystal structure of Human FancF Protein that Functions in the Assembly of a DNA Damage Signaling Complex | | Descriptor: | Fanconi anemia group F protein, MERCURY (II) ION | | Authors: | Kowal, P, Gurtan, A.M, Stuckert, P, Lehmann, C, D'Andrea, A, Ellenberger, T.E. | | Deposit date: | 2006-10-13 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural determinants of human FANCF protein that function in the assembly of a DNA damage signaling complex.
J.Biol.Chem., 282, 2007
|
|
2O0J
 
 | | T4 gp17 ATPase domain mutant complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA packaging protein Gp17 | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2006-11-27 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the ATPase that Powers DNA Packaging into Bacteriophage T4 Procapsids
MOL.CELL, 25, 2007
|
|
2KTT
 
 | | Solution Structure of a Covalently Bound Pyrrolo[2,1-c][1,4]benzodiazepine-Benzimidazole Hybrid to a 10mer DNA Duplex | | Descriptor: | (11aS)-7-methoxy-8-(3-{4-[6-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]phenoxy}propoxy)-1,2,3,10,11,11a-hexahydro-5H-pyrrolo[2,1-c][1,4]benzodiazepin-5-one, 5'-D(*AP*AP*CP*AP*AP*TP*TP*GP*TP*T)-3' | | Authors: | Rettig, M, Weingarth, M, Langel, W, Kamal, A, Kumar, P.P, Weisz, K. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a covalently bound pyrrolo[2,1-c][1,4]benzodiazepine-benzimidazole hybrid to a 10mer DNA duplex.
Biochemistry, 48, 2009
|
|
8R64
 
 | |
7P2N
 
 | | E.coli GyrB24 with inhibitor LSJ38 (EBL2684) | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-5-oxidanyl-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Exploring the 5-Substituted 2-Aminobenzothiazole-Based DNA Gyrase B Inhibitors Active against ESKAPE Pathogens.
Acs Omega, 8, 2023
|
|
7X9F
 
 | | Crystal structure of actinomycin D-echinomycin-d(AGCGCGT/ACGCGCT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(P*AP*CP*GP*CP*GP*CP*T)-3'), DNA (5'-D(P*AP*GP*CP*GP*CP*GP*T)-3'), ... | | Authors: | Kao, S.H, Satange, R.B, Hou, M.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.
Nucleic Acids Res., 50, 2022
|
|
7X6R
 
 | | Crystal structure of actinomycin D-echinomycin-d(AGCACGT/ACGGGCT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, Actinomycin D, COBALT (II) ION, ... | | Authors: | Kao, S.H, Satange, R.B, Hou, M.H. | | Deposit date: | 2022-03-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.
Nucleic Acids Res., 50, 2022
|
|
7X97
 
 | | Crystal structure of actinomycin D-echinomycin-d(AGCCCGT/ACGGGCT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, Actinomycin D, CHLORIDE ION, ... | | Authors: | Kao, S.H, Satange, R.B, Hou, M.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.
Nucleic Acids Res., 50, 2022
|
|
7XDJ
 
 | | Crystal structure of actinomycin D-echinomycin-d(AGCGCGT/ACGAGCT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, CHLORIDE ION, DNA (5'-D(P*AP*CP*GP*AP*GP*CP*(BRU))-3'), ... | | Authors: | Chien, C.M, Satange, R.B, Hou, M.H. | | Deposit date: | 2022-03-27 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.
Nucleic Acids Res., 50, 2022
|
|
3QG5
 
 | |
3HSL
 
 | | The Crystal Structure of PF-8, the DNA Polymerase Accessory Subunit from Kaposi's Sarcoma-Associated Herpesvirus | | Descriptor: | ORF59 | | Authors: | Baltz, J.L, Filman, D.J, Ciustea, M, Silverman, J.E.Y, Lautenschlager, C.L, Coen, D.M, Ricciardi, R.P, Hogle, J.M. | | Deposit date: | 2009-06-10 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of PF-8, the DNA polymerase accessory subunit from Kaposi's sarcoma-associated herpesvirus.
J.Virol., 83, 2009
|
|
7XG2
 
 | | CryoEM structure of type IV-A NTS-nicked dsDNA bound Csf-crRNA ternary complex | | Descriptor: | Csf1, Csf2, Csf3, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-04-02 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Type IV-A CRISPR-Csf complex: Assembly, dsDNA targeting, and CasDinG recruitment.
Mol.Cell, 83, 2023
|
|
3I5E
 
 | |
3I5L
 
 | | Allosteric Modulation of DNA by Small Molecules | | Descriptor: | (22R,51R)-22,51-diamino-5,11,17,28,34,40,46,57-octamethyl-2,5,8,11,14,17,20,25,28,31,34,37,40,43,46,49,54,57,60,61,64,6 5-docosaazanonacyclo[54.2.1.1~4,7~.1~10,13~.1~16,19~.1~27,30~.1~33,36~.1~39,42~.1~45,48~]hexahexaconta-1(58),4(66),6,10( 65),12,16(64),18,27(63),29,33(62),35,39(61),41,45(60),47,56(59)-hexadecaene-3,9,15,21,26,32,38,44,50,55-decone, 5'-D(*CP*CP*AP*GP*GP*(C38)P*CP*TP*GP*G)-3', CALCIUM ION | | Authors: | Chenoweth, D.M, Dervan, P.B. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Allosteric modulation of DNA by small molecules
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KWQ
 
 | | Mcm10 C-terminal DNA binding domain | | Descriptor: | Protein MCM10 homolog, ZINC ION | | Authors: | Robertson, P.D, Chagot, B, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the C-terminal DNA binding domain of Mcm10 reveals a conserved MCM motif.
J.Biol.Chem., 285, 2010
|
|
3I2M
 
 | | The Crystal Structure of PF-8, the DNA Polymerase Accessory Subunit from Kaposi s Sarcoma-Associated Herpesvirus | | Descriptor: | ORF59 | | Authors: | Baltz, J.L, Filman, D.J, Ciustea, M, Silverman, J.E.Y, Lautenschlager, C.L, Coen, D.M, Ricciardi, R.P, Hogle, J.M. | | Deposit date: | 2009-06-29 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The crystal structure of PF-8, the DNA polymerase accessory subunit from Kaposi's sarcoma-associated herpesvirus.
J.Virol., 83, 2009
|
|
2VS7
 
 | | The crystal structure of I-DmoI in complex with DNA and Ca | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP *CP*TP*TP*AP*CP*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP *GP*TP*AP*AP*GP*TP*TP*CP*CP*GP*GP*CP*GP*CP*G)-3', ACETATE ION, ... | | Authors: | Marcaida, M.J, Prieto, J, Redondo, P, Nadra, A.D, Alibes, A, Serrano, L, Grizot, S, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2008-04-21 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of I-Dmoi in Complex with its Target DNA Provides New Insights Into Meganuclease Engineering.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3WRS
 
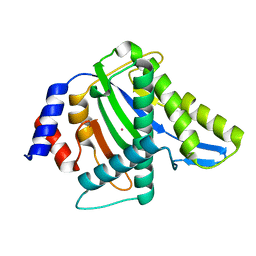 | |
3WRQ
 
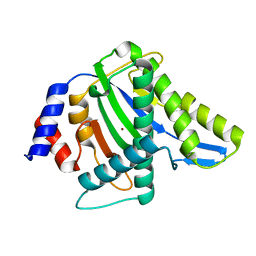 | |
8SKI
 
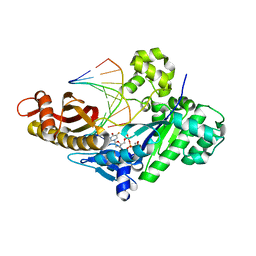 | |
7P2W
 
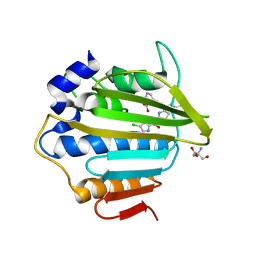 | | E.coli GyrB24 with inhibitor LMD92 (EBL2682) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(3-carboxyphenyl)methoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, ... | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7P2M
 
 | | E.coli GyrB24 with inhibitor LMD43 (EBL2560) | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-phenylmethoxy-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
5Z3V
 
 | | Structure of Snf2-nucleosome complex at shl-2 in ADP BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5Z3O
 
 | | Structure of Snf2-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (167-MER), Histone H2A, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
