7RDS
 
 | | Structure of human NTHL1 | | Descriptor: | IRON/SULFUR CLUSTER, Isoform 3 of Endonuclease III-like protein 1 | | Authors: | Carroll, B.L, Zahn, K.E, Doublie, S. | | Deposit date: | 2021-07-11 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Caught in motion: human NTHL1 undergoes interdomain rearrangement necessary for catalysis.
Nucleic Acids Res., 49, 2021
|
|
7RDT
 
 | | Structure of human NTHL1 - linker 1 chimera | | Descriptor: | IRON/SULFUR CLUSTER, Isoform 3 of Endonuclease III-like protein 1 | | Authors: | Carroll, B.L, Doublie, S. | | Deposit date: | 2021-07-11 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Caught in motion: human NTHL1 undergoes interdomain rearrangement necessary for catalysis.
Nucleic Acids Res., 49, 2021
|
|
2IMP
 
 | | Crystal structure of lactaldehyde dehydrogenase from E. coli: the ternary complex with lactate (occupancy 0.5) and NADH. Crystals soaked with (L)-Lactate. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, LACTIC ACID, Lactaldehyde dehydrogenase, ... | | Authors: | Di Costanzo, L, Gomez, G.A, Christianson, D.W. | | Deposit date: | 2006-10-04 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Lactaldehyde Dehydrogenase from Escherichia coli and Inferences Regarding Substrate and Cofactor Specificity.
J.Mol.Biol., 366, 2007
|
|
7ROJ
 
 | | Amyloid-related segment of alphaB-crystallin residues 90-100 with G95W mutation | | Descriptor: | Alpha-crystallin B chain peptide, GLYCEROL, SODIUM ION, ... | | Authors: | Sawaya, M.R, Do, T.D, Eisenberg, D.S. | | Deposit date: | 2021-07-30 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic view of an amyloid dodecamer exhibiting selective cellular toxic vulnerability in acute brain slices.
Protein Sci., 31, 2022
|
|
7RJ6
 
 | | CRYSTAL STRUCTURE OF AP2 ASSOCIATED KINASE 1 ISOFORM 1 COMPLEXED WITH LIGAND (2R)-2-AMINO-N-[3-(DIFLUOROM ETHOXY)-4-(1,3-OXAZOL-5-YL)PHENYL]-4-METHYLPENTANAMIDE | | Descriptor: | (1S)-1-[4-ethyl-6-(1,3-oxazol-5-yl)quinazolin-2-yl]-3-methylbutan-1-amine, 5-[(4-aminopiperidin-1-yl)methyl]-N-{3-[5-(propan-2-yl)-1,3,4-thiadiazol-2-yl]phenyl}pyrrolo[2,1-f][1,2,4]triazin-4-amine, AP2-associated protein kinase 1, ... | | Authors: | Muckelbauer, J. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.132 Å) | | Cite: | Bicyclic Heterocyclic Replacement of an Aryl Amide Leading to Potent and Kinase-Selective Adaptor Protein 2-Associated Kinase 1 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RJ7
 
 | |
5JAC
 
 | | Sixty minutes iron loaded Rana Catesbeiana H' ferritin variant E57A/E136A/D140A | | Descriptor: | CHLORIDE ION, FE (II) ION, Ferritin, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S, Bernacchioni, C, Turano, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Ferroxidase Activity in Eukaryotic Ferritin is Controlled by Accessory-Iron-Binding Sites in the Catalytic Cavity.
Chemistry, 22, 2016
|
|
3I5X
 
 | | Structure of Mss116p bound to ssRNA and AMP-PNP | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase MSS116, MAGNESIUM ION, ... | | Authors: | Del Campo, M, Lambowitz, A.M. | | Deposit date: | 2009-07-06 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA
Mol.Cell, 35, 2009
|
|
6NFW
 
 | |
5J9V
 
 | | Ten minutes iron loaded Rana Catesbeiana H' ferritin variant E57A/E136A/D140A | | Descriptor: | CHLORIDE ION, FE (II) ION, Ferritin, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S, Bernacchioni, C, Turano, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Ferroxidase Activity in Eukaryotic Ferritin is Controlled by Accessory-Iron-Binding Sites in the Catalytic Cavity.
Chemistry, 22, 2016
|
|
1HKU
 
 | | CtBP/BARS: a dual-function protein involved in transcription corepression and Golgi membrane fission | | Descriptor: | C-TERMINAL BINDING PROTEIN 3, FORMIC ACID, GLYCEROL, ... | | Authors: | Nardini, M, Spano, S, Cericola, C, Pesce, A, Massaro, A, Millo, E, Luini, A, Corda, D, Bolognesi, M. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-19 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ctbp/Bars: A Dual-Function Protein Involved in Transcription Co-Repression and Golgi Membrane Fission
Embo J., 22, 2003
|
|
1HLB
 
 | |
6E9E
 
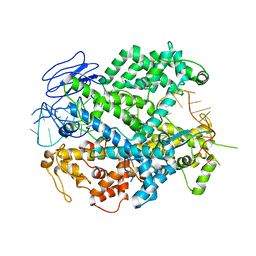 | | EsCas13d-crRNA binary complex | | Descriptor: | EsCas13d, MAGNESIUM ION, crRNA (52-MER) | | Authors: | Zhang, C, Lyumkis, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d.
Cell, 175, 2018
|
|
1NR4
 
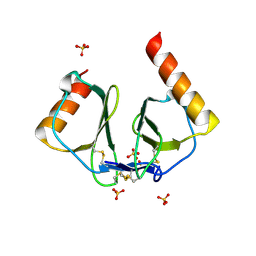 | | High resolution crystal structures of thymus and activation-regulated chemokine | | Descriptor: | SULFATE ION, Thymus and activation-regulated chemokine | | Authors: | Asojo, O.A, Boulegue, C, Hoover, D.M, Lu, W, Lubkowski, J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-08-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structures of thymus and activation-regulated chemokine (TARC).
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1K5C
 
 | | Endopolygalacturonase I from Stereum purpureum at 0.96 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ENDOPOLYGALACTURONASE, ... | | Authors: | Shimizu, T, Nakatsu, T, Miyairi, K, Okuno, T, Kato, H. | | Deposit date: | 2001-10-10 | | Release date: | 2002-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution.
Biochemistry, 41, 2002
|
|
1NR2
 
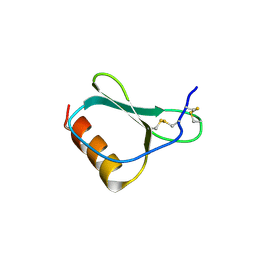 | | High resolution crystal structures of thymus and activation-regulated chemokine | | Descriptor: | Thymus and activation-regulated chemokine | | Authors: | Asojo, O.A, Boulegue, C, Hoover, D.M, Lu, W, Lubkowski, J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-08-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of thymus and activation-regulated chemokine (TARC).
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5J8S
 
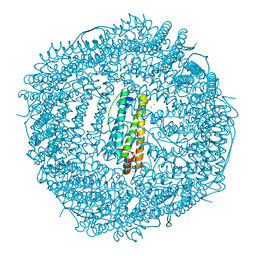 | | Iron-free state of Rana Catesbeiana H' ferritin variant E57A/E136A/D140A | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S, Bernacchioni, C, Turano, P. | | Deposit date: | 2016-04-08 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ferroxidase Activity in Eukaryotic Ferritin is Controlled by Accessory-Iron-Binding Sites in the Catalytic Cavity.
Chemistry, 22, 2016
|
|
7SHG
 
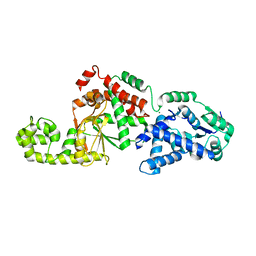 | |
6RPU
 
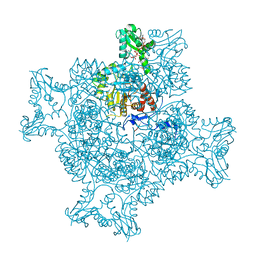 | | Structure of the ternary complex of the IMPDH enzyme from Ashbya gossypii bound to the dinucleoside polyphosphate Ap5G and GDP | | Descriptor: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Buey, R.M, Fernandez-Justel, D, Revuelta, J.L. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Bateman domain of IMP dehydrogenase is a binding target for dinucleoside polyphosphates.
J.Biol.Chem., 294, 2019
|
|
8UPP
 
 | | Campylobacter jejuni ketol-acid reductoisomerase in complex with NADPH and Hoe704 | | Descriptor: | (2R)-(dimethylphosphoryl)(hydroxy)acetic acid, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Lin, X, Lv, Y, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2023-10-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mapping of the Reaction Trajectory catalyzed by Class I Ketol-Acid Reductoisomerase
Acs Catalysis, 2024
|
|
8UPN
 
 | |
8UPQ
 
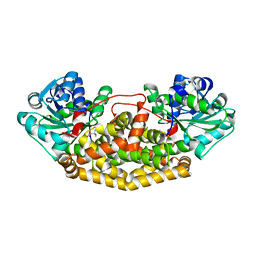 | | Campylobacter jejuni ketol-acid reductoisomerase in complex with 2,3-dihydroxy-3-isovalerate. | | Descriptor: | (2R)-2,3-dihydroxy-3-methylbutanoic acid, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION | | Authors: | Lin, X, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2023-10-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mapping of the Reaction Trajectory catalyzed by Class I Ketol-Acid Reductoisomerase
Acs Catalysis, 2024
|
|
1KCD
 
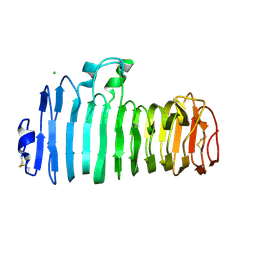 | | Endopolygalacturonase I from Stereum purpureum complexed with two galacturonate at 1.15 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ENDOPOLYGALACTURONASE, ... | | Authors: | Shimizu, T, Nakatsu, T, Miyairi, K, Okuno, T, Kato, H. | | Deposit date: | 2001-11-08 | | Release date: | 2002-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution.
Biochemistry, 41, 2002
|
|
1KCC
 
 | | Endopolygalacturonase I from Stereum purpureum complexed with a galacturonate at 1.00 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ENDOPOLYGALACTURONASE, ... | | Authors: | Shimizu, T, Nakatsu, T, Miyairi, K, Okuno, T, Kato, H. | | Deposit date: | 2001-11-08 | | Release date: | 2002-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution.
Biochemistry, 41, 2002
|
|
9END
 
 | | Crystal structure of Methanopyrus kandleri malate dehydrogenase mutant 3 | | Descriptor: | CHLORIDE ION, Malate dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Coquille, S, Roche, J, Girard, E, Madern, D. | | Deposit date: | 2024-03-12 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Navigating the conformational landscape of an enzyme. Stabilization of a low populated conformer by evolutionary mutations triggers Allostery into a non-allosteric enzyme.
To be published
|
|
