7CHO
 
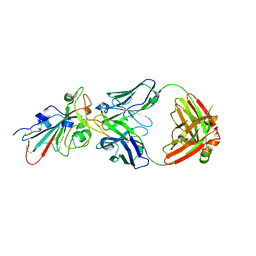 | | Crystal structure of SARS-CoV-2 antibody P5A-1D2 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-1D2 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
8TFQ
 
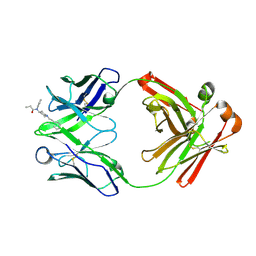 | | Fab from C10-S66K antibody in complex with fentanyl | | Descriptor: | Heavy chain from Fab of C10_S66K antibody, Light chain from Fab of C10_S66K antibody, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide | | Authors: | Pholcharee, T, Wilson, I.A. | | Deposit date: | 2023-07-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Engineered Human-Antibody Fragment with Fentanyl Pan-Specificity That Reverses Carfentanil-Induced Respiratory Depression.
Acs Chem Neurosci, 14, 2023
|
|
4UZF
 
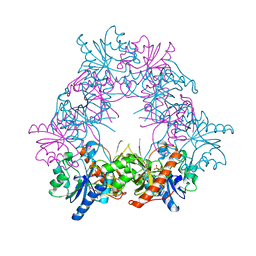 | |
7CK8
 
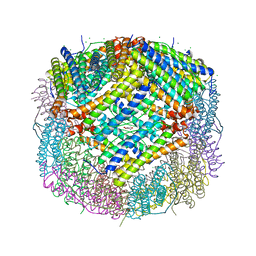 | | Crystal structure of human ferritin heavy chain mutant C90S/C102S/C130S | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin heavy chain, ... | | Authors: | Chen, X, Jiang, B, Yan, X, Fan, K. | | Deposit date: | 2020-07-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A natural drug entry channel in the ferritin nanocage.
Nano Today, 35, 2020
|
|
4U7F
 
 | | Reduced quinone reductase 2 in complex with CK2 inhibitor DMAT | | Descriptor: | 4,5,6,7-TETRABROMO-N,N-DIMETHYL-1H-BENZIMIDAZOL-2-AMINE, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Leung, K.K, Shilton, B.H. | | Deposit date: | 2014-07-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Quinone Reductase 2 Is an Adventitious Target of Protein Kinase CK2 Inhibitors TBBz (TBI) and DMAT.
Biochemistry, 54, 2015
|
|
4ZP8
 
 | | Coxsackievirus B3 Polymerase - F364L mutant | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Peersen, O.B, McDonald, S.M. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Design of a Genetically Stable High Fidelity Coxsackievirus B3 Polymerase That Attenuates Virus Growth in Vivo.
J.Biol.Chem., 291, 2016
|
|
4TZ3
 
 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
1EOQ
 
 | | ROUS SARCOMA VIRUS CAPSID PROTEIN: C-TERMINAL DOMAIN | | Descriptor: | GAG POLYPROTEIN CAPSID PROTEIN P27 | | Authors: | Kingston, R.L, Fitzon-Ostendorp, T, Eisenmesser, E.Z, Schatz, G.W, Vogt, V.M, Post, C.B, Rossmann, M.G. | | Deposit date: | 2000-03-23 | | Release date: | 2000-08-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and self-association of the Rous sarcoma virus capsid protein.
Structure Fold.Des., 8, 2000
|
|
9B9N
 
 | | Crystal structure of FlcD from Pseudomonas aeruginosa bound to iron (II) and substrate | | Descriptor: | (2R)-2-{[(2Z)-2-(hydroxyimino)ethyl]sulfanyl}butanedioic acid, FE (III) ION, Pyrroloquinoline quinone (Coenzyme PQQ) biosynthesis protein C | | Authors: | Walker, M.E, Grove, T.L, Li, B, Redinbo, M.R. | | Deposit date: | 2024-04-02 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis for Methine Excision by a Heme Oxygenase-like Enzyme
Acs Cent.Sci., 2024
|
|
9B9M
 
 | |
9BNR
 
 | | 4-(2-isothiocyanatoethyl)benzenesulfonamide complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | Macrophage migration inhibitory factor, N-[2-(4-sulfamoylphenyl)ethyl]methanethioamide, SULFATE ION | | Authors: | Fellner, M, Rutledge, M.T, Putha, L, Kok, L.K, Gamble, A.B, Wilbanks, S.M, Vernall, A.J, Tyndall, J.D.A. | | Deposit date: | 2024-05-02 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Covalent isothiocyanate inhibitors of macrophage migration inhibitory factor as potential colorectal cancer treatments.
Chemmedchem, 2024
|
|
1EMZ
 
 | | SOLUTION STRUCTURE OF FRAGMENT (350-370) OF THE TRANSMEMBRANE DOMAIN OF HEPATITIS C ENVELOPE GLYCOPROTEIN E1 | | Descriptor: | ENVELOPE GLYCOPROTEIN E1 | | Authors: | Op De Beeck, A, Montserret, R, Duvet, S, Cocquerel, L, Cacan, R, Barberot, B, Le Maire, M, Penin, F, Dubuisson, J. | | Deposit date: | 2000-03-20 | | Release date: | 2000-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization.
J.Biol.Chem., 275, 2000
|
|
4ZOU
 
 | |
4ZPA
 
 | | Coxsackievirus B3 Polymerase - F364Y mutant | | Descriptor: | RNA-directed RNA polymerase, SULFATE ION | | Authors: | Peersen, O.B, McDonald, S.M. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.665 Å) | | Cite: | Design of a Genetically Stable High Fidelity Coxsackievirus B3 Polymerase That Attenuates Virus Growth in Vivo.
J.Biol.Chem., 291, 2016
|
|
4ZP6
 
 | | Coxsackievirus B3 Polymerase - F364A mutant | | Descriptor: | Genome polyprotein, SULFATE ION | | Authors: | Peersen, O.B, McDonald, S.M. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Design of a Genetically Stable High Fidelity Coxsackievirus B3 Polymerase That Attenuates Virus Growth in Vivo.
J.Biol.Chem., 291, 2016
|
|
4U6S
 
 | | CtBP1 in complex with substrate phenylpyruvate | | Descriptor: | 3-PHENYLPYRUVIC ACID, C-terminal-binding protein 1, CALCIUM ION, ... | | Authors: | Hilbert, B.J, Morris, B.L, Ellis, K.C, Paulsen, J.L, Schiffer, C.A, Grossman, S.R, Royer Jr, W.E. | | Deposit date: | 2014-07-29 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of a High Affinity Inhibitor to Human CtBP.
Acs Chem.Biol., 10, 2015
|
|
4ZP9
 
 | | Coxsackievirus B3 Polymerase - F364I mutant | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Peersen, O.B, McDonald, S.M. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Design of a Genetically Stable High Fidelity Coxsackievirus B3 Polymerase That Attenuates Virus Growth in Vivo.
J.Biol.Chem., 291, 2016
|
|
7CBG
 
 | | Crystal structure of threonyl-tRNA synthetase (ThrRS) from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-N-[(E)-4-[6,7-bis(chloranyl)-4-oxidanylidene-quinazolin-3-yl]but-2-enyl]-2-(methylamino)-3-oxidanyl-butanamide, Threonine--tRNA ligase, ZINC ION | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2020-06-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-guided optimization and mechanistic study of a class of quinazolinone-threonine hybrids as antibacterial ThrRS inhibitors.
Eur.J.Med.Chem., 207, 2020
|
|
4ZUJ
 
 | |
4ZW1
 
 | | Crystal structure of hBRD4 in complex with BL-BI06 reveals a novel synthesized inhibitor that induces Beclin1-independent/ATG5-dependent autophagic cell death in breast cancer | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-hydroxy-3,5-dimethylphenyl)-7-methyl-5,6,7,8-tetrahydropyrido[4',3':4,5]thieno[2,3-d]pyrimidin-4(3H)-one, Bromodomain-containing protein 4 | | Authors: | Liu, B, Zhang, S, Guo, M, Tian, M. | | Deposit date: | 2015-05-19 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of hBRD4 in complex with BL-BI06 reveals a novel synthesized inhibitor that induces Beclin1-independent/ATG5-dependent autophagic cell death in breast cancer
To Be Published
|
|
4ZWI
 
 | | Surface Lysine Acetylated Human Carbonic Anhydrase II in Complex with a Sulfamate-Based Inhibitor | | Descriptor: | (6R)-1-O-acetyl-2,6-anhydro-6-{[4-(sulfamoyloxy)piperidin-1-yl]sulfonyl}-L-glucitol, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Lomelino, C.L, Mahon, B.P, McKenna, M. | | Deposit date: | 2015-05-19 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Observed surface lysine acetylation of human carbonic anhydrase II expressed in Escherichia coli.
Protein Sci., 24, 2015
|
|
4ZWZ
 
 | | Engineered Carbonic Anhydrase IX mimic in complex with a glucosyl sulfamate inhibitor | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ZINC ION, ... | | Authors: | Mahon, B.P, Lomelino, C.L, Driscoll, J.M, McKenna, R. | | Deposit date: | 2015-05-19 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mapping Selective Inhibition of the Cancer-Related Carbonic Anhydrase IX Using Structure-Activity Relationships of Glucosyl-Based Sulfamates.
J.Med.Chem., 58, 2015
|
|
4U7H
 
 | | Oxidized quinone reductase 2 in complex with CK2 inhibitor DMAT | | Descriptor: | 4,5,6,7-TETRABROMO-N,N-DIMETHYL-1H-BENZIMIDAZOL-2-AMINE, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Leung, K.K, Shilton, B.H. | | Deposit date: | 2014-07-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Quinone Reductase 2 Is an Adventitious Target of Protein Kinase CK2 Inhibitors TBBz (TBI) and DMAT.
Biochemistry, 54, 2015
|
|
5A5L
 
 | | Structure of dual function FBPase SBPase from Thermosynechococcus elongatus | | Descriptor: | 7-O-phosphono-alpha-L-galacto-hept-2-ulopyranose, D-FRUCTOSE 1,6-BISPHOSPHATASE CLASS 2/SEDOHEPTULOSE 1,7-BISPHOSPHATASE, MAGNESIUM ION, ... | | Authors: | Cotton, C.A.R, Kabasakal, B, Miah, N, Murray, J.W. | | Deposit date: | 2015-06-19 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of the Dual-Function Fructose-1,6/Sedoheptulose-1, 7-Bisphosphatase from Thermosynechococcus Elongatus Bound with Sedoheptulose-7-Phosphate.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4ZSB
 
 | |
