3V4W
 
 | | Structure of E347K mutant of Lamin | | Descriptor: | Prelamin-A/C | | Authors: | Bollati, M, Bolognesi, M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structures of the lamin A/C R335W and E347K mutants: Implications for dilated cardiolaminopathies.
Biochem.Biophys.Res.Commun., 418, 2012
|
|
2DNP
 
 | | Solution structure of RNA binding domain 2 in RNA-binding protein 14 | | Descriptor: | RNA-binding protein 14 | | Authors: | Kusuhara, M, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain 2 in RNA-binding protein 14
To be Published
|
|
7R1D
 
 | | Structure of MuvB complex | | Descriptor: | Histone-binding protein RBBP4, Protein lin-37 homolog, Protein lin-9 homolog | | Authors: | Koliopoulos, M.G, Alfieri, C. | | Deposit date: | 2022-02-02 | | Release date: | 2022-08-10 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a nucleosome-bound MuvB transcription factor complex reveals DNA remodelling.
Nat Commun, 13, 2022
|
|
7SGS
 
 | | Cryo-EM structure of full-length MAP7 bound to the microtubule | | Descriptor: | Ensconsin, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ferro, L.S, Fang, Q, Eshun-Wilson, L, Fernandes, J, Jack, A, Farrell, D.P, Golcuk, M, Huijben, T, Costa, K, Gur, M, DiMaio, F, Nogales, E, Yildiz, A. | | Deposit date: | 2021-10-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional insight into regulation of kinesin-1 by microtubule-associated protein MAP7.
Science, 375, 2022
|
|
7S6H
 
 | | Human PARP1 deltaV687-E688 bound to NAD+ analog EB-47 and to a DNA double strand break. | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, DNA (5'-D(*CP*GP*AP*CP*G)-3'), ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-14 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
7S81
 
 | | Structure of human PARP1 domains (Zn1, Zn3, WGR, HD) bound to a DNA double strand break. | | Descriptor: | DNA (5'-D(*AP*TP*GP*CP*GP*GP*CP*CP*GP*CP*AP*T)-3'), Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
7S6M
 
 | |
7S68
 
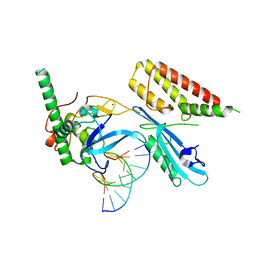 | | Structure of human PARP1 domains (Zn1, Zn3, WGR and HD) bound to a DNA double strand break. | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*CP*AP*GP*GP*C)-3'), Fusion of PARP1 zinc fingers 1 and 3 (Zn1, Zn3), ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
3VHX
 
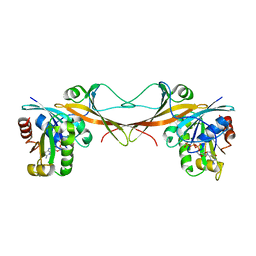 | | The crystal structure of Arf6-MKLP1 (Mitotic kinesin-like protein 1) complex | | Descriptor: | ADP-ribosylation factor 6, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Makyio, H, Takei, T, Ohgi, H, Takahashi, S, Takatsu, H, Ueda, T, Kanaho, Y, Xie, Y, Shin, H.W, Kamikubo, H, Kataoka, M, Kawasaki, M, Kato, R, Wakatsuki, S, Nakayama, K. | | Deposit date: | 2011-09-12 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for Arf6-MKLP1 complex formation on the Flemming body responsible for cytokinesis
Embo J., 31, 2012
|
|
5LQF
 
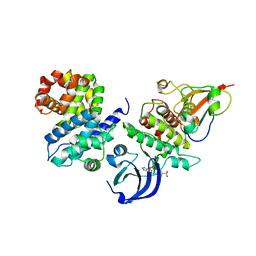 | | CDK1/CyclinB1/CKS2 in complex with NU6102 | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B.J, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E, Newell, D.R, Turner, D.M, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Endicott, J.A, Cano, C. | | Deposit date: | 2016-08-17 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
3W99
 
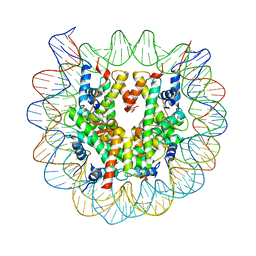 | | Crystal Structure of Human Nucleosome Core Particle lacking H4 N-terminal region | | Descriptor: | 146-mer DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Iwasaki, W, Miya, Y, Horikoshi, N, Osakabe, A, Tachiwana, H, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2013-04-01 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Contribution of histone N-terminal tails to the structure and stability of nucleosomes
FEBS Open Bio, 3, 2013
|
|
7SUD
 
 | | CryoEM structure of DNA-PK complex VIII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, MAGNESIUM ION, ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
3WA9
 
 | | The nucleosome containing human H2A.Z.1 | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B type 1-J, ... | | Authors: | Horikoshi, N, Sato, K, Shimada, K, Arimura, Y, Osakabe, A, Tachiwana, H, Iwasaki, W, Kagawa, W, Harata, M, Kimura, H, Kurumizaka, H. | | Deposit date: | 2013-04-30 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural polymorphism in the L1 loop regions of human H2A.Z.1 and H2A.Z.2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3W96
 
 | | Crystal Structure of Human Nucleosome Core Particle lacking H2A N-terminal region | | Descriptor: | 146-mer DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Miya, Y, Horikoshi, N, Osakabe, A, Tachiwana, H, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2013-04-01 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Contribution of histone N-terminal tails to the structure and stability of nucleosomes
FEBS Open Bio, 3, 2013
|
|
3WAA
 
 | | The nucleosome containing human H2A.Z.2 | | Descriptor: | DNA (146-MER), Histone H2A.V, Histone H2B type 1-J, ... | | Authors: | Horikoshi, N, Sato, K, Shimada, K, Arimura, Y, Osakabe, A, Tachiwana, H, Iwasaki, W, Kagawa, W, Harata, M, Kimura, H, Kurumizaka, H. | | Deposit date: | 2013-04-30 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural polymorphism in the L1 loop regions of human H2A.Z.1 and H2A.Z.2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7U0G
 
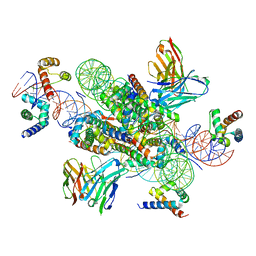 | | structure of LIN28b nucleosome bound 3 OCT4 | | Descriptor: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Lian, T, Guan, R, Bai, Y. | | Deposit date: | 2022-02-18 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
7U0J
 
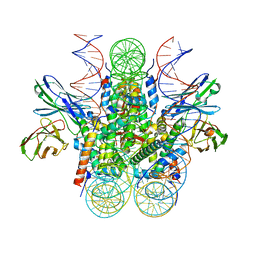 | | Structure of 162bp LIN28b nucleosome | | Descriptor: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Lian, T, Guan, R, Bai, Y. | | Deposit date: | 2022-02-18 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
7U0I
 
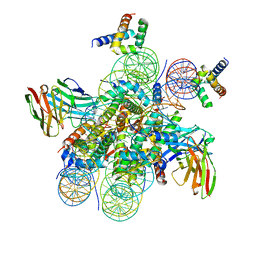 | | Structure of LIN28b nucleosome bound 2 OCT4 | | Descriptor: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Tengfei, L, Guan, R, Bai, Y. | | Deposit date: | 2022-02-18 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
5MR8
 
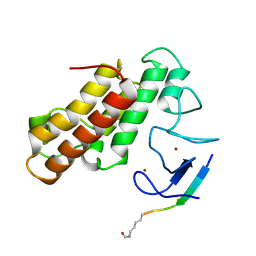 | | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K9ac histone peptide | | Descriptor: | E3 ubiquitin-protein ligase TRIM33, Histone H3, ZINC ION | | Authors: | Tallant, C, Savitsky, P, Fedorov, O, Nunez-Alonso, G, Siejka, P, Krojer, T, Williams, E, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K9ac histone peptide
To Be Published
|
|
2BF8
 
 | | Crystal structure of SUMO modified ubiquitin conjugating enzyme E2- 25K | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2-25 KDA, UBIQUITIN-LIKE PROTEIN SMT3C | | Authors: | Pichler, A, Knipscheer, P, Oberhofer, E, Van Dijk, W.J, Korner, R, Velgaard Olsen, J, Jentsch, S, Melchior, F, Sixma, T.K. | | Deposit date: | 2004-12-06 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sumo Imodification of the Ubiquitin Conjugating Enzyme E2-25K
Nat.Struct.Mol.Biol., 12, 2005
|
|
7SYX
 
 | | Structure of the delta dII IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S24, 40S ribosomal protein S25, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYW
 
 | | Structure of the wt IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(wt) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7U46
 
 | |
7U47
 
 | |
2CQG
 
 | | Solution structure of the RNA binding domain of TAR DNA-binding protein-43 | | Descriptor: | TAR DNA-binding protein-43 | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain of TAR DNA-binding protein-43
To be Published
|
|
