7B84
 
 | | Notum-Fragment065 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B8C
 
 | | Notum-Fragment 147 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methyl-~{N}-(2-methylpropyl)imidazo[1,2-a]pyridine-3-carboxamide, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B87
 
 | | Notum-Fragment074 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B86
 
 | | Notum-Fragment067 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(1~{H}-pyrazol-3-yl)phenoxy]pyrimidine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B8D
 
 | | Notum-Fragment 151 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-amino-N-(pyridin-2-yl)benzenesulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
6WU6
 
 | | succinate-coenzyme Q reductase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Lyu, M, Su, C.-C, Morgan, C.E, Yu, E.W. | | Deposit date: | 2020-05-04 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
7B89
 
 | | Notum-Fragment077 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7SVH
 
 | | Bile Salt Hydrolase B from Lactobacillus gasseri | | Descriptor: | Choloylglycine hydrolase, MAGNESIUM ION | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2021-11-19 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Bile salt hydrolases shape the bile acid landscape and restrict Clostridioides difficile growth in the murine gut.
Nat Microbiol, 8, 2023
|
|
7SVJ
 
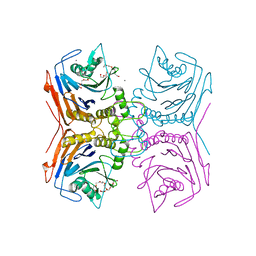 | | Bile Salt Hydrolase from Lactobacillus ingluviei | | Descriptor: | CALCIUM ION, Choloylglycine hydrolase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Walker, M.E, Patel, S, Redinbo, M.R. | | Deposit date: | 2021-11-19 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Bile salt hydrolases shape the bile acid landscape and restrict Clostridioides difficile growth in the murine gut.
Nat Microbiol, 8, 2023
|
|
6MII
 
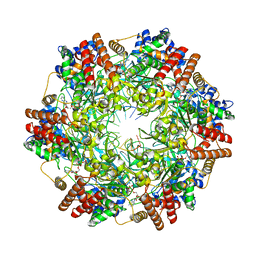 | | Crystal structure of minichromosome maintenance protein MCM/DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Enemark, E.J, Meagher, M, Epling, L.B. | | Deposit date: | 2018-09-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | DNA translocation mechanism of the MCM complex and implications for replication initiation.
Nat Commun, 10, 2019
|
|
7SVI
 
 | |
7SVG
 
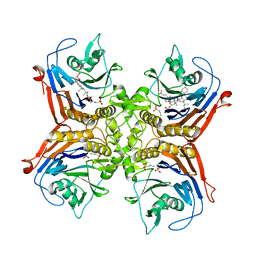 | |
7Q5E
 
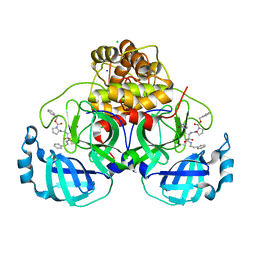 | | Crystal structure of F2F-2020209-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2021-11-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Easy access to alpha-ketoamides as SARS-CoV-2 and MERS M pro inhibitors via the PADAM oxidation route.
Eur.J.Med.Chem., 244, 2022
|
|
7Q5F
 
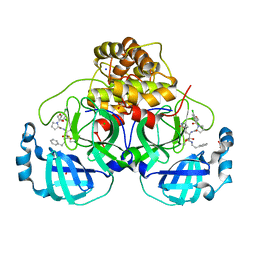 | | Crystal structure of F2F-2020216-01X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (S)-1-(2-(2,4-dichlorophenoxy)acetyl)-N-((S)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)-4-(phenethylamino)butan-2-yl)pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, 3C-like proteinase, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2021-11-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Easy access to alpha-ketoamides as SARS-CoV-2 and MERS M pro inhibitors via the PADAM oxidation route.
Eur.J.Med.Chem., 244, 2022
|
|
7SVK
 
 | |
5LVP
 
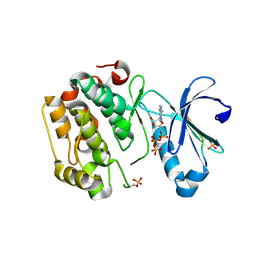 | | Human PDK1 Kinase Domain in Complex with an HM-Peptide Bound to the PIF-Pocket | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schulze, J.O, Saladino, G, Busschots, K, Neimanis, S, Suess, E, Odadzic, D, Zeuzem, S, Hindie, V, Herbrand, A.K, Lisa, M.N, Alzari, P.M, Gervasio, F.L, Biondi, R.M. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-19 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bidirectional Allosteric Communication between the ATP-Binding Site and the Regulatory PIF Pocket in PDK1 Protein Kinase.
Cell Chem Biol, 23, 2016
|
|
7B3N
 
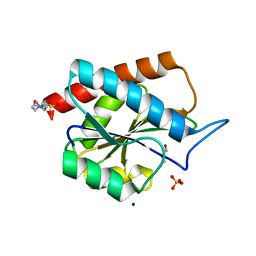 | | AmiP amidase-3 from Thermus parvatiensis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Cell wall hydrolase, ... | | Authors: | Freitag-Pohl, S, Pohl, E. | | Deposit date: | 2020-12-01 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | AmiP from hyperthermophilic Thermus parvatiensis prophage is a thermoactive and ultrathermostable peptidoglycan lytic amidase.
Protein Sci., 32, 2023
|
|
7SVF
 
 | |
7SVE
 
 | |
7T8S
 
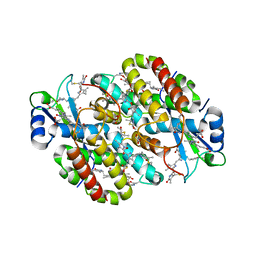 | | Light Harvesting complex phycoerythrin PE 566, from the cryptophyte Cryptomonas pyrenoidifera | | Descriptor: | Bilin 584 (doubly linked), Bilin 584 (single linked), Bilin 618 (single linked), ... | | Authors: | Michie, K.A, Harrop, S.J, Rathbone, H.W, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2021-12-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structures reveal the origin of spectral variation in cryptophyte light harvesting antenna proteins.
Protein Sci., 32, 2023
|
|
6PB1
 
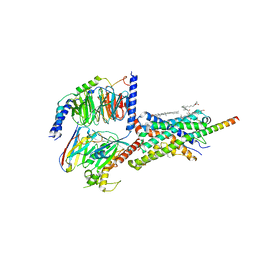 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 2 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
6WDS
 
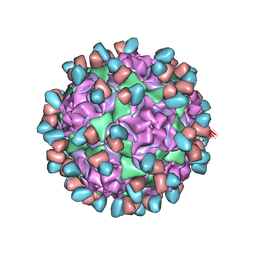 | | Enterovirus D68 in complex with human monoclonal antibody EV68-159 | | Descriptor: | EV68-159 heavy chain, EV68-159 light chain, viral protein 1, ... | | Authors: | Fu, J, Klose, T, Vogt, M.R, Crowe, J.E, Rossmann, M.G, Kuhn, R.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human antibodies neutralize enterovirus D68 and protect against infection and paralytic disease.
Sci Immunol, 5, 2020
|
|
8DK6
 
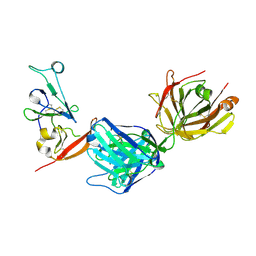 | | Structure of hepatitis C virus envelope N-terminal truncated glycoprotein 2 (E2) (residues 456-713) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, 2A12 Fab light chain, ... | | Authors: | Kumar, A, Rohe, T, Elrod, E.J, Khan, A.G, Dearborn, A.D, Kissinger, R, Grakoui, A, Marcotrigiano, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Regions of hepatitis C virus E2 required for membrane association.
Nat Commun, 14, 2023
|
|
6AN8
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 8.0 | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA PRIMER (5'- D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*GP)-3'), DNA TEMPLATE (5'- D(*AP*TP*GP*AP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-08-12 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
6ASW
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 9.0 | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA (27-MER), DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DDG))-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-08-25 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
