4U9S
 
 | | Crystal structure of NqrC from Vibrio cholerae | | Descriptor: | BROMIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Fritz, G. | | Deposit date: | 2014-08-06 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase.
Nature, 516, 2014
|
|
4UAJ
 
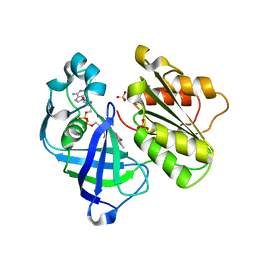 | | Crystal structure of NqrF in hexagonal space group | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit F, SULFATE ION | | Authors: | Fritz, G. | | Deposit date: | 2014-08-10 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7019 Å) | | Cite: | Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase.
Nature, 516, 2014
|
|
4MM3
 
 | |
6XA4
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW241 | | Descriptor: | 3C-like proteinase, GLYCEROL, inhibitor UAW241 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
1MVO
 
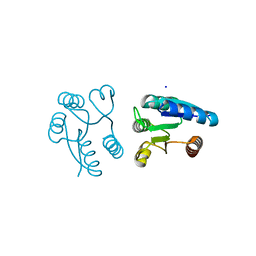 | | Crystal structure of the PhoP receiver domain from Bacillus subtilis | | Descriptor: | MANGANESE (II) ION, PhoP response regulator, SODIUM ION | | Authors: | Birck, C, Chen, Y, Hulett, F.M, Samama, J.P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-09-26 | | Release date: | 2002-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Phosphorylation Domain in PhoP Reveals a Functional Tandem Association Mediated by an Asymmetric Interface
J.BACTERIOL., 185, 2003
|
|
6XLQ
 
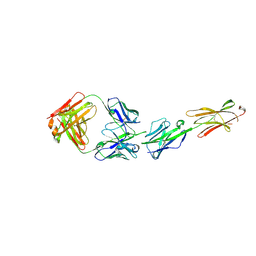 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the CTX-2026 Fab | | Descriptor: | Butyrophilin subfamily 3 member A1, CTX-2026 Heavy Chain, CTX-2026 Light Chain | | Authors: | Payne, K.K, Mine, J.A, Biswas, S, Chaurio, R.A, Perales-Puchalt, A, Anadon, C.M, Costich, T.L, Harro, C.M, Walrath, J, Ming, Q, Tcyganov, E, Buras, A.L, Rigolizzo, K.E, Mandal, G, Lajoie, J, Ophir, M, Tchou, J, Marchion, D, Luca, V.C, Bobrowicz, P, McLaughlin, B, Eskiocak, U, Schmidt, M, Cubillos-Ruiz, J.R, Rodriguez, P.C, Gabrilovich, D.I, Conejo-Garcia, J.R. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | BTN3A1 governs antitumor responses by coordinating alpha beta and gamma delta T cells.
Science, 369, 2020
|
|
1N37
 
 | |
5GGX
 
 | |
4WPE
 
 | | Crystal Structure of Hof1p F-BAR domain | | Descriptor: | Cytokinesis protein 2 | | Authors: | Lemmon, M.A, Moravcevic, K. | | Deposit date: | 2014-10-17 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of Saccharomyces cerevisiae F-BAR Domain Structures Reveals a Conserved Inositol Phosphate Binding Site.
Structure, 23, 2015
|
|
7BBV
 
 | | Pectate lyase B from Verticillium dahliae | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Pectate lyase B, ... | | Authors: | Safran, J, Habrylo, O, Bouckaert, J, Pau Roblot, C, Senechal, F, Pelloux, J. | | Deposit date: | 2020-12-18 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The specificity of pectate lyase VdPelB from Verticilium dahliae is highlighted by structural, dynamical and biochemical characterizations.
Int.J.Biol.Macromol., 231, 2023
|
|
7CV2
 
 | | Crystal structure of B. halodurans NiaR in niacin-bound form | | Descriptor: | NICOTINIC ACID, Transcriptional regulator NiaR, ZINC ION | | Authors: | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
5GGY
 
 | |
2NUX
 
 | |
5GVZ
 
 | |
2NUW
 
 | |
4YS4
 
 | | Crystal structure of Pf41 tandem 6-cys domains from Plasmodium falciparum | | Descriptor: | CHLORIDE ION, GLYCEROL, Merozoite surface protein P41, ... | | Authors: | Parker, M.L, Peng, F, Boulanger, M.J. | | Deposit date: | 2015-03-16 | | Release date: | 2015-10-14 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Structure of Plasmodium falciparum Blood-Stage 6-Cys Protein Pf41 Reveals an Unexpected Intra-Domain Insertion Required for Pf12 Coordination.
Plos One, 10, 2015
|
|
2PVS
 
 | | Structure of human pancreatic lipase related protein 2 mutant N336Q | | Descriptor: | CALCIUM ION, Pancreatic lipase-related protein 2, SULFATE ION | | Authors: | Spinelli, S, Eydoux, C, Carriere, F, Cambillau, C. | | Deposit date: | 2007-05-10 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human pancreatic lipase-related protein 2 with the lid in an open conformation.
Biochemistry, 47, 2008
|
|
7CV0
 
 | | Crystal structure of B. halodurans NiaR in apo form | | Descriptor: | Transcriptional regulator NiaR, ZINC ION | | Authors: | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
4Z7X
 
 | | MdbA protein, a thiol-disulfide oxidoreductase from Actinomyces oris. | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, MdbA | | Authors: | OSIPIUK, J, Reardon-Robinson, M.E, Ton-That, H, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-04-08 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Disulfide Bond-forming Machine Is Linked to the Sortase-mediated Pilus Assembly Pathway in the Gram-positive Bacterium Actinomyces oris.
J.Biol.Chem., 290, 2015
|
|
4ZE3
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) Y140H mutant complexed with fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R, Monk, B.C, Tyndall, J.D.A. | | Deposit date: | 2015-04-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14 alpha-demethylase.
Sci Rep, 6, 2016
|
|
4WWC
 
 | | Crystal structure of full length YvoA in complex with palindromic operator DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*CP*AP*GP*TP*GP*GP*TP*CP*TP*AP*GP*AP*CP*CP*AP*CP*TP*GP*G)-3'), HTH-type transcriptional repressor YvoA | | Authors: | Grau, F.C, Fillenberg, S.B, Muller, Y.A. | | Deposit date: | 2014-11-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structural insight into operator dre-sites recognition and effector binding in the GntR/HutC transcription regulator NagR.
Nucleic Acids Res., 43, 2015
|
|
4YAS
 
 | | HYDROXYNITRILE LYASE COMPLEXED WITH CHLORALHYDRATE | | Descriptor: | PROTEIN (HYDROXYNITRILE LYASE), SULFATE ION, TRI-CHLORO-ACETALDEHYDE | | Authors: | Zuegg, J, Wagner, U.G, Gugganig, M, Kratky, C. | | Deposit date: | 1999-03-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of enzyme-substrate complexes of the hydroxynitrile lyase from Hevea brasiliensis.
Protein Sci., 8, 1999
|
|
2PGZ
 
 | | Crystal structure of Cocaine bound to an ACh-Binding Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COCAINE, Soluble acetylcholine receptor, ... | | Authors: | Hansen, S.B, Taylor, P. | | Deposit date: | 2007-04-10 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Galanthamine and non-competitive inhibitor binding to ACh-binding protein: evidence for a binding site on non-alpha-subunit interfaces of heteromeric neuronal nicotinic receptors.
J.Mol.Biol., 369, 2007
|
|
3HA6
 
 | | Crystal structure of aurora A in complex with TPX2 and compound 10 | | Descriptor: | N~2~-(3,4-dimethoxyphenyl)-N~4~-[2-(2-fluorophenyl)ethyl]-N~6~-quinolin-6-yl-1,3,5-triazine-2,4,6-triamine, Serine/threonine-protein kinase 6, Targeting protein for Xklp2 | | Authors: | Zhao, B, Clark, M.A. | | Deposit date: | 2009-05-01 | | Release date: | 2009-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Design, synthesis and selection of DNA-encoded small-molecule libraries.
Nat.Chem.Biol., 5, 2009
|
|
2PO3
 
 | | Crystal Structure Analysis of DesI in the presence of its TDP-sugar product | | Descriptor: | (2R,3R,4S,5S,6R)-3,4-DIHYDROXY-5-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)IMINO]-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL [(2R,3S,5R)-3-HYDROXY-5-(5-METHYL-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, 4-dehydrase | | Authors: | Burgie, E.S, Holden, H.M. | | Deposit date: | 2007-04-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Architecture of DesI: A Key Enzyme in the Biosynthesis of Desosamine
Biochemistry, 46, 2007
|
|
