4B9O
 
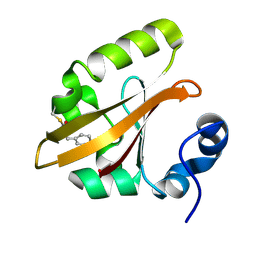 | | The PR0 Photocycle Intermediate of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Schotte, F, Cho, H.S, Kaila, V.R.I, Kamikubo, H, Dashdorj, N, Henry, E.R, Graber, T.J, Henning, R, Wulff, M, Hummer, G, Kataoka, M, Anfinrud, P.A. | | Deposit date: | 2012-09-06 | | Release date: | 2012-11-14 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching a Signaling Protein Function in Real Time Via 100-Ps Time-Resolved Laue Crystallography
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4B2Q
 
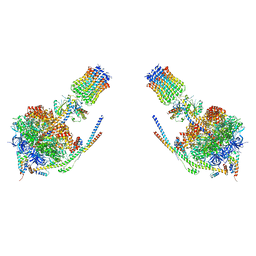 | |
6URJ
 
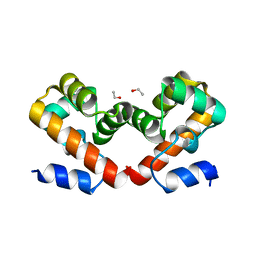 | | Barrier-to-autointegration factor soaked in Acetone: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6USB
 
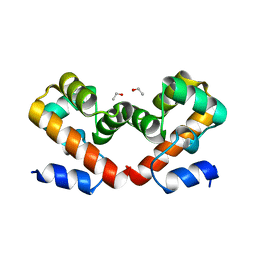 | | Barrier-to-autointegration factor soaked in urea: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6UW2
 
 | | Clotrimazole bound complex of Acanthamoeba castellanii CYP51 | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, GUANIDINE, Obtusifoliol 14alphademethylase, ... | | Authors: | Sharma, V, Podust, L.M. | | Deposit date: | 2019-11-04 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Domain-Swap Dimerization of Acanthamoeba castellanii CYP51 and a Unique Mechanism of Inactivation by Isavuconazole.
Mol.Pharmacol., 98, 2020
|
|
6US1
 
 | | Barrier-to-autointegration factor soaked in isobutanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6US7
 
 | | Barrier-to-autointegration factor soaked in TMAO: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3PRU
 
 | | Crystal Structure of Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, rod 1 (fragment 14-158) from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR182A | | Descriptor: | CHLORIDE ION, Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, ... | | Authors: | Kuzin, A, Su, M, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-30 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR182A
To be Published
|
|
4ABF
 
 | | Fragments bound to bovine trypsin for the SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 1-(5-BROMO-1-BENZOTHIOPHEN-3-YL)METHANAMINE, CALCIUM ION, ... | | Authors: | Newman, J, Peat, T.S. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Dingo Dataset: A Comprehensive Set of Data for the Sampl Challenge.
J.Comput.Aided Mol.Des., 26, 2012
|
|
6URL
 
 | | Barrier-to-autointegration factor soaked in isopropanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URR
 
 | | Barrier-to-autointegration factor soaked in Dioxane: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6USI
 
 | | Barrier-to-autointegration factor soaked in 1,6-hexanediol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4ABG
 
 | | Fragments bound to bovine trypsin for the SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-(4-METHYLPIPERAZIN-1-YL)PHENYL]METHANAMINE, CALCIUM ION, ... | | Authors: | Newman, J, Peat, T.S. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The Dingo Dataset: A Comprehensive Set of Data for the Sampl Challenge.
J.Comput.Aided Mol.Des., 26, 2012
|
|
6UX0
 
 | | Isavuconazole bound complex of Acanthamoeba castellanii CYP51 | | Descriptor: | 4-{2-[(2R,3R)-3-(2,5-difluorophenyl)-3-hydroxy-4-(1H-1,2,4-triazol-1-yl)butan-2-yl]-1,3-thiazol-4-yl}benzonitrile, FE (III) ION, Obtusifoliol 14alphademethylase, ... | | Authors: | Sharma, V, Podust, L.M. | | Deposit date: | 2019-11-06 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Domain-Swap Dimerization of Acanthamoeba castellanii CYP51 and a Unique Mechanism of Inactivation by Isavuconazole.
Mol.Pharmacol., 98, 2020
|
|
8G1W
 
 | | Crystal Structure Matriptase (C731S) in Complex with Inhibitor VD4162B | | Descriptor: | CHLORIDE ION, Cyclic peptide inhibitor (ACE)Y(DTR)(NLE)(KCM), Cyclic peptide inhibitor (ACE)Y(DTR)(NLE)(THZ), ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Janetka, J.W. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mechanism-Based Macrocyclic Inhibitors of Serine Proteases.
J.Med.Chem., 67, 2024
|
|
6URZ
 
 | | Barrier-to-autointegration factor soaked in methanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL, METHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URN
 
 | | Barrier-to-autointegration factor t-butanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6US0
 
 | | Barrier-to-autointegration factor soaked in R,S,R-bisfuranol (RSR): 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8G1V
 
 | | Crystal Structure Matriptase (C731S) in Complex with Inhibitor MM1132-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, N~2~-{[3-(acetamidomethyl)phenyl]acetyl}-N-[(2S)-1-(1,3-benzothiazol-2-yl)-5-carbamimidamido-1,1-dihydroxypentan-2-yl]-L-leucinamide, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Janetka, J.W. | | Deposit date: | 2023-02-03 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Use of protease substrate specificity screening in the rational design of selective protease inhibitors with unnatural amino acids: Application to HGFA, matriptase, and hepsin.
Protein Sci., 33, 2024
|
|
4BBT
 
 | | The PR1 Photocycle Intermediate of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Schotte, F, Cho, H.S, Kaila, V.R.I, Kamikubo, H, Dashdorj, N, Henry, E.R, Graber, T.J, Henning, R, Wulff, M, Hummer, G, Kataoka, M, Anfinrud, P.A. | | Deposit date: | 2012-09-27 | | Release date: | 2012-11-14 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching a Signaling Protein Function in Real Time Via 100-Ps Time-Resolved Laue Crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8F2K
 
 | |
6W62
 
 | | Cryo-EM structure of Cas12i-crRNA complex | | Descriptor: | Cas12i, crRNA | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms for target recognition and cleavage by the Cas12i RNA-guided endonuclease.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4ABI
 
 | | Co-complex structure of bovine trypsin with a modified Bowman-Birk inhibitor (PtA)SFTI-1(1,14), that was 1,4-disubstituted with a 1,2,3- trizol to mimic a trans amide bond | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, DIMETHYLFORMAMIDE, ... | | Authors: | Schmelz, S, Empting, M, Tischler, M, Nasu, D, Heinz, D, Kolmar, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Braces for the Peptide Backbone: Insights Into Structure-Activity Relation-Ships of Protease Inhibitor Mimics with Locked Amide Conformations
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4ABJ
 
 | | Co-complex structure of bovine trypsin with a modified Bowman-Birk inhibitor (IcA)SFTI-1(1,14), that was 1,5-disubstituted with 1,2,3- trizol to mimic a cis amide bond | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, DIMETHYLFORMAMIDE, ... | | Authors: | Schmelz, S, Empting, M, Tischler, M, Nasu, D, Heinz, D, Kolmar, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Braces for the Peptide Backbone: Insights Into Structure-Activity Relation-Ships of Protease Inhibitor Mimics with Locked Amide Conformations
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3QRL
 
 | |
