7UJT
 
 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B(S1303D) in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type II subunit alpha, GLYCEROL, ... | | Authors: | Ozden, C, Stratton, M.M, Garman, S.C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
7UJS
 
 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type II subunit alpha, Glutamate receptor ionotropic, ... | | Authors: | Ozden, C, Santos, N.J, Stratton, M.M, Garman, S.C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
7UJR
 
 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B | | Descriptor: | 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase type II subunit alpha, Glutamate receptor ionotropic, ... | | Authors: | Ozden, C, Santos, N.J, Stratton, M.M, Garman, S.C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
7UJQ
 
 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha, GLYCEROL, Glutamate receptor ionotropic, ... | | Authors: | Ozden, C, Foster, J.C, Stratton, M.M, Garman, S.C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
7UJP
 
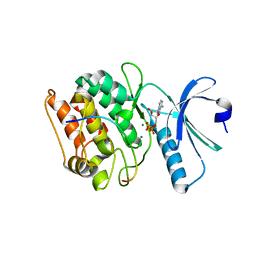 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type II subunit alpha, ... | | Authors: | Ozden, C, Foster, J.C, Stratton, M.M, Garman, S.C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
2VNK
 
 | |
8T9N
 
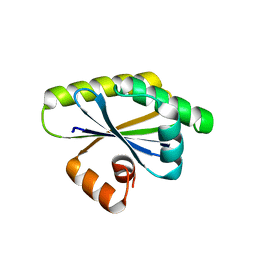 | | Bacillus subtilis RsgI GGG mutant | | Descriptor: | Anti-sigma-I factor RsgI | | Authors: | Brogan, A.P, Habib, C, Hobbs, S.J, Kranzusch, P.J, Rudner, D.Z. | | Deposit date: | 2023-06-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacterial SEAL domains undergo autoproteolysis and function in regulated intramembrane proteolysis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2VF0
 
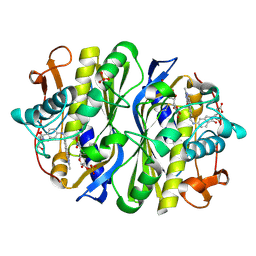 | | CRYSTAL STRUCTURE OF THE THYMIDYLATE SYNTHASE K48Q COMPLEXED WITH 5NO2DUMP AND BW1843U89 | | Descriptor: | 2'-DEOXY-5-NITROURIDINE 5'-MONOPHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, SULFATE ION, ... | | Authors: | Sotelo-Mundo, R.R, Arreola, R, Maley, F, Montfort, W.R. | | Deposit date: | 2007-10-27 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Role of an Invariant Lysine Residue in Folate Binding on Escherichia Coli Thymidylate Synthase: Calorimetric and Crystallographic Analysis of the K48Q Mutant.
Int.J.Biochem.Cell Biol., 40, 2008
|
|
7NEZ
 
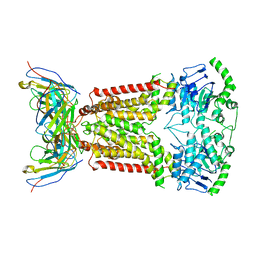 | | Structure of topotecan-bound ABCG2 | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, ... | | Authors: | Kowal, J, Locher, K, Ni, D, Stahlberg, H. | | Deposit date: | 2021-02-05 | | Release date: | 2021-04-21 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural Basis of Drug Recognition by the Multidrug Transporter ABCG2.
J.Mol.Biol., 433, 2021
|
|
7NFD
 
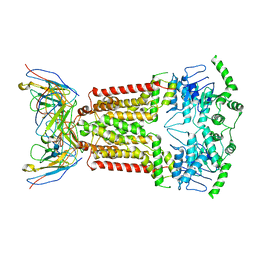 | | Structure of mitoxantrone-bound ABCG2 | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, ... | | Authors: | Kowal, J, Locher, K, Ni, D, Stahlberg, H. | | Deposit date: | 2021-02-06 | | Release date: | 2021-04-21 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural Basis of Drug Recognition by the Multidrug Transporter ABCG2.
J.Mol.Biol., 433, 2021
|
|
7NEQ
 
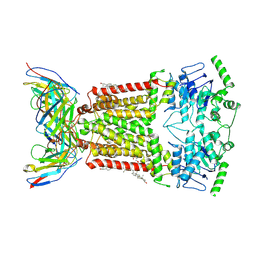 | | Structure of tariquidar-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Kowal, J, Locher, K. | | Deposit date: | 2021-02-04 | | Release date: | 2021-04-21 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural Basis of Drug Recognition by the Multidrug Transporter ABCG2.
J.Mol.Biol., 433, 2021
|
|
6EST
 
 | |
8OK0
 
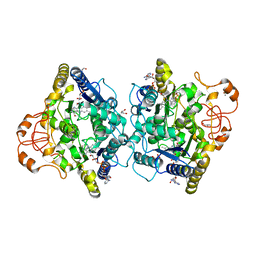 | | Crystal structure of human NQO1 in complex with the inhibitor PMSF | | Descriptor: | ACETYL GROUP, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Martin-Garcia, J.M, Grieco, A, Ruiz-Fresneda, M.A. | | Deposit date: | 2023-03-26 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural dynamics at the active site of the cancer-associated flavoenzyme NQO1 probed by chemical modification with PMSF.
Febs Lett., 597, 2023
|
|
2L78
 
 | |
2PNR
 
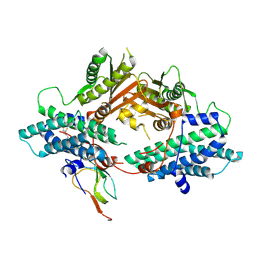 | | Crystal Structure of the asymmetric Pdk3-l2 Complex | | Descriptor: | DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 3 | | Authors: | Vassylyev, D.G, Steussy, C.N, Devedjiev, Y. | | Deposit date: | 2007-04-25 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Asymmetric complex of Pyruvate Dehydrogenase
Kinase 3 with Lipoyl domain 2 and its Biological Implications
J.Mol.Biol., 370, 2007
|
|
6MK8
 
 | |
8TA3
 
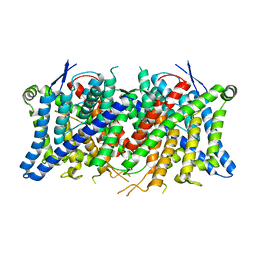 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain Apo state with resolved N-terminal hairpin | | Descriptor: | CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8SO5
 
 | |
1NFO
 
 | | APOLIPOPROTEIN E2 (APOE2, D154A MUTATION) | | Descriptor: | APOLIPOPROTEIN E2 | | Authors: | Rupp, B, Parkin, S. | | Deposit date: | 1996-07-17 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia.
Nat.Struct.Biol., 3, 1996
|
|
1NH4
 
 | |
7FAJ
 
 | | CARM1 bound with compound 43 | | Descriptor: | Histone-arginine methyltransferase CARM1, N'-[[3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)-5-methyl-6-phenylazanyl-pyrimidin-2-yl]phenyl]methyl]-N-methyl-ethane-1,2-diamine | | Authors: | Cao, D.Y, Li, J, Xiong, B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2450726 Å) | | Cite: | Structure-Based Discovery of Potent CARM1 Inhibitors for Solid Tumor and Cancer Immunology Therapy.
J.Med.Chem., 64, 2021
|
|
3HZW
 
 | | Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog bothropstoxin-1, p-Bromophenacyl bromide | | Authors: | Fernandes, C.A.H, Marchi-Salvador, D.P, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
1ODB
 
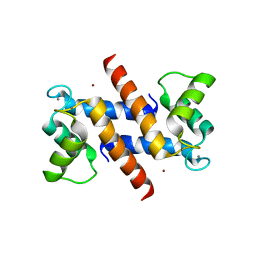 | | THE CRYSTAL STRUCTURE OF HUMAN S100A12 - COPPER COMPLEX | | Descriptor: | CALCIUM ION, CALGRANULIN C, COPPER (II) ION | | Authors: | Moroz, O.V, Antson, A.A, Grist, S.J, Maitland, N.J, Dodson, G.G, Wilson, K.S, Lukanidin, E.M, Bronstein, I.B. | | Deposit date: | 2003-02-15 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the Human S100A12-Copper Complex: Implications for Host-Parasite Defence
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6GSS
 
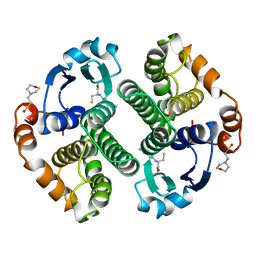 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
8U95
 
 | | The structure of myosin heavy chain from Drosophila melanogaster flight muscle thick filaments | | Descriptor: | Myosin heavy chain, isoform U | | Authors: | Abbasi Yeganeh, F, Rastegarpouyani, H, Li, J, Taylor, K.A. | | Deposit date: | 2023-09-18 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the Drosophila melanogaster Flight Muscle Myosin Filament at 4.7 angstrom Resolution Reveals New Details of Non-Myosin Proteins.
Int J Mol Sci, 24, 2023
|
|
