2A48
 
 | | Crystal structure of amFP486 E150Q | | Descriptor: | BETA-MERCAPTOETHANOL, GFP-like fluorescent chromoprotein amFP486 | | Authors: | Henderson, J.N, Remington, S.J. | | Deposit date: | 2005-06-28 | | Release date: | 2005-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and mutational analysis of amFP486, a cyan fluorescent protein from Anemonia majano
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1QXT
 
 | | Crystal structure of precyclized intermediate for the green fluorescent protein R96A variant (A) | | Descriptor: | green-fluorescent protein | | Authors: | Barondeau, D.P, Putnam, C.D, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2003-09-08 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures
Proc.Natl.Acad.Sci.USA, 100
|
|
8B6T
 
 | | X-ray structure of the interface optimized haloalkane dehalogenase HaloTag7 fusion to the green fluorescent protein GFP (ChemoG5-TMR) labeled with a chloroalkane tetramethylrhodamine fluorophore substrate | | Descriptor: | CHLORIDE ION, Green fluorescent protein,Haloalkane dehalogenase, [9-[2-carboxy-5-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]phenyl]-6-(dimethylamino)xanthen-3-ylidene]-dimethyl-azanium | | Authors: | Tarnawski, M, Hellweg, L, Hiblot, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A general method for the development of multicolor biosensors with large dynamic ranges.
Nat.Chem.Biol., 19, 2023
|
|
1QY3
 
 | | Crystal structure of precyclized intermediate for the green fluorescent protein R96A variant (B) | | Descriptor: | green-fluorescent protein | | Authors: | Barondeau, D.P, Putnam, C.D, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2003-09-09 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
4OWJ
 
 | |
7A8G
 
 | |
7A7V
 
 | |
7A7Q
 
 | | rsGreenF-K206A in the green-on state | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Green fluorescent protein | | Authors: | De Zitter, E, Dedecker, P, Van Meervelt, L. | | Deposit date: | 2020-08-30 | | Release date: | 2021-02-17 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Function Dataset Reveals Environment Effects within a Fluorescent Protein Model System*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7A8I
 
 | |
4P0M
 
 | | Crystal structure of an evolved putative penicillin-binding protein homolog, Rv2911, from Mycobacterium tuberculosis | | Descriptor: | D-alanyl-D-alanine carboxypeptidase | | Authors: | Krieger, I, Yu, M, Bursey, E, Hung, L.-W, Terwilliger, T.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subfamily-Specific Adaptations in the Structures of Two Penicillin-Binding Proteins from Mycobacterium tuberculosis.
Plos One, 9, 2014
|
|
5WD6
 
 | | bovine salivary protein form 30b | | Descriptor: | CALCIUM ION, Short palate, lung and nasal epithelium carcinoma-associated protein 2B | | Authors: | Zhang, H, Arcus, V.L. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-11 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three dimensional structure of Bovine Salivary Protein 30b (BSP30b) and its interaction with specific rumen bacteria.
Plos One, 14, 2019
|
|
4PPK
 
 | | Crystal structure of eCGP123 T69V variant at pH 7.5 | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
4PPR
 
 | | Crystal structure of Mycobacterium tuberculosis D,D-peptidase Rv3330 in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein DacB1 | | Authors: | Prigozhin, D.M, Huizar, J.P, Mavrici, D, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-02-27 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subfamily-specific adaptations in the structures of two penicillin-binding proteins from Mycobacterium tuberculosis.
Plos One, 9, 2014
|
|
1EMG
 
 | | GREEN FLUORESCENT PROTEIN (65-67 REPLACED BY CRO, S65T SUBSTITUTION, Q80R) | | Descriptor: | PROTEIN (GREEN FLUORESCENT PROTEIN) | | Authors: | Elsliger, M.A, Wachter, R.M, Kallio, K, Hanson, G.T, Remington, S.J. | | Deposit date: | 1998-11-12 | | Release date: | 1999-05-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and spectral response of green fluorescent protein variants to changes in pH.
Biochemistry, 38, 1999
|
|
2EMD
 
 | |
3UR2
 
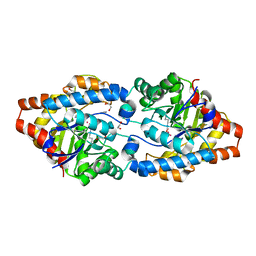 | | Crystal Structure of PTE mutant H254G/H257W/L303T/K185R/I274N/A80V | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, IMIDAZOLE, ... | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
1GP1
 
 | |
3HPQ
 
 | |
7YV3
 
 | |
4TF4
 
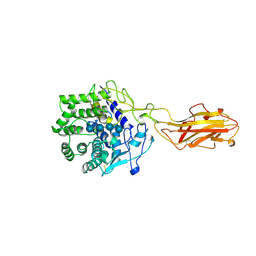 | | ENDO/EXOCELLULASE:CELLOPENTAOSE FROM THERMOMONOSPORA | | Descriptor: | CALCIUM ION, T. FUSCA ENDO/EXO-CELLULASE E4 CATALYTIC DOMAIN AND CELLULOSE-BINDING DOMAIN, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Sakon, J, Wilson, D.B, Karplus, P.A. | | Deposit date: | 1997-05-31 | | Release date: | 1997-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca.
Nat.Struct.Biol., 4, 1997
|
|
1Z1P
 
 | |
6QFK
 
 | | EngBF DARPin Fusion 4b G10 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
1AKE
 
 | |
2Q57
 
 | |
8F8F
 
 | | The structure of Rv2173 from M. tuberculosis (APO form) | | Descriptor: | (2E,6E)-farnesyl diphosphate synthase, GLYCEROL, MAGNESIUM ION | | Authors: | Johnston, J.M, Allison, T.M, Titterington, J. | | Deposit date: | 2022-11-22 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of Rv2173 from M. tuberculosis in APO-, IPP-, and DMAP-bound forms.
To be Published
|
|
