4O9K
 
 | | Crystal structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Methylococcus capsulatus in complex with CMP-Kdo | | Descriptor: | Arabinose 5-phosphate isomerase, CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, GLYCEROL | | Authors: | Shabalin, I.G, Cooper, D.R, Shumilin, I.A, Zimmerman, M.D, Majorek, K.A, Hammonds, J, Hillerich, B.S, Nawar, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and kinetic properties of D-arabinose 5-phosphate isomerase from Methylococcus capsulatus
To be Published
|
|
4O9X
 
 | | Crystal Structure of TcdB2-TccC3 | | Descriptor: | MERCURY (II) ION, TcdB2, TccC3 | | Authors: | Meusch, D, Gatsogiannis, C, Efremov, R.G, Lang, A.E, Hofnagel, O, Vetter, I.R, Aktories, K, Raunser, S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Mechanism of Tc toxin action revealed in molecular detail.
Nature, 508, 2014
|
|
3EIU
 
 | | A second transient position of ATP on its trail to the nucleotide-binding site of subunit B of the motor protein A1Ao ATP synthase | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ADENOSINE-5'-TRIPHOSPHATE, V-type ATP synthase beta chain | | Authors: | Manimekalai, S.M.S, Kumar, A, Balakrishna, A.M, Gruber, G. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | A second transient position of ATP on its trail to the nucleotide-binding site of subunit B of the motor protein A(1)A(O) ATP synthase
J.Struct.Biol., 166, 2009
|
|
3E3K
 
 | | Structural characterization of a putative endogenous metal chelator in the periplasmic nickel transporter NikA (butane-1,2,4-tricarboxylate without nickel form) | | Descriptor: | (2R)-butane-1,2,4-tricarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Cherrier, M.V, Cavazza, C, Bochot, C, Lemaire, D, Fontecilla-Camps, J.C. | | Deposit date: | 2008-08-07 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of a putative endogenous metal chelator in the periplasmic nickel transporter NikA
Biochemistry, 47, 2008
|
|
3V5X
 
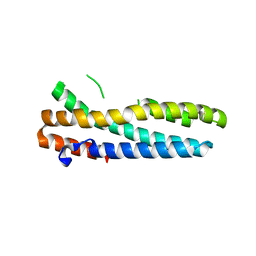 | | Structure of FBXL5 hemerythrin domain, C2 cell | | Descriptor: | F-box/LRR-repeat protein 5, MU-OXO-DIIRON | | Authors: | Tomchick, D.R, Bruick, R.K, Thompson, J.W, Brautigam, C.A. | | Deposit date: | 2011-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Molecular Characterization of Iron-sensing Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5).
J.Biol.Chem., 287, 2012
|
|
3V9O
 
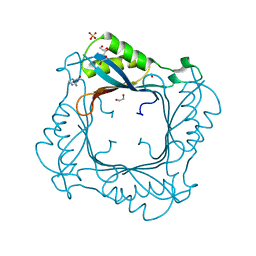 | |
3E57
 
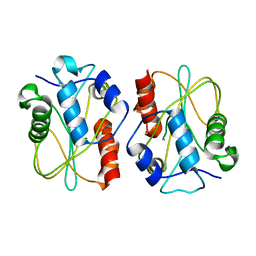 | |
4OGF
 
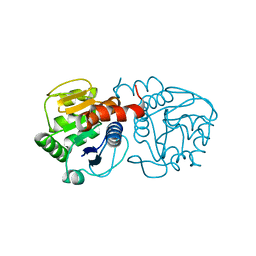 | |
3E5T
 
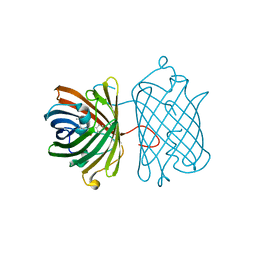 | | Crystal Structure Analysis of FP611 | | Descriptor: | Red fluorescent protein eqFP611 | | Authors: | Nar, H, Nienhaus, K, Nienhaus, U, Wiedenmann, J. | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Trans-cis isomerization is responsible for the red-shifted fluorescence in variants of the red fluorescent protein eqFP611.
J.Am.Chem.Soc., 130, 2008
|
|
3EMM
 
 | | X-ray structure of protein from Arabidopsis thaliana AT1G79260 with Bound Heme | | Descriptor: | 1,2-ETHANEDIOL, PROTOPORPHYRIN IX CONTAINING FE, Uncharacterized protein At1g79260 | | Authors: | Bianchetti, C.M, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-09-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.358 Å) | | Cite: | The structure and NO binding properties of the nitrophorin-like heme-binding protein from Arabidopsis thaliana gene locus At1g79260.1.
Proteins, 78, 2010
|
|
3E7C
 
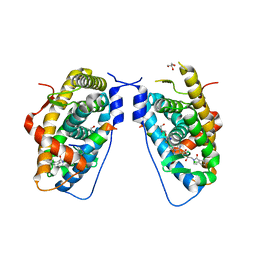 | | Glucocorticoid Receptor LBD bound to GSK866 | | Descriptor: | 5-amino-N-[(2S)-2-({[(2,6-dichlorophenyl)carbonyl](ethyl)amino}methyl)-3,3,3-trifluoro-2-hydroxypropyl]-1-(4-fluorophenyl)-1H-pyrazole-4-carboxamide, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Madauss, K.P, Williams, S.P, Mclay, I, Stewart, E.L, Bledsoe, R.K. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The first X-ray crystal structure of the glucocorticoid receptor bound to a non-steroidal agonist.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3E3P
 
 | |
3V9K
 
 | |
3VF1
 
 | | Structure of a calcium-dependent 11R-lipoxygenase suggests a mechanism for Ca-regulation | | Descriptor: | 11R-lipoxygenase, FE (II) ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Eek, P, Jarving, R, Jarving, I, Gilbert, N.C, Newcomer, M.E, Samel, N. | | Deposit date: | 2012-01-09 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Structure of a Calcium-dependent 11R-Lipoxygenase Suggests a Mechanism for Ca2+ Regulation.
J.Biol.Chem., 287, 2012
|
|
3E86
 
 | |
3E4B
 
 | | Crystal structure of AlgK from Pseudomonas fluorescens WCS374r | | Descriptor: | AlgK, CHLORIDE ION, GLYCEROL | | Authors: | Keiski, C.-L, Harwich, M, Jain, S, Neculai, A.M, Yip, P, Robinson, H, Whitney, J.C, Burrows, L.L, Ohman, D.E, Howell, P.L. | | Deposit date: | 2008-08-11 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin.
Structure, 18, 2010
|
|
4OLR
 
 | | [Leu-5]-Enkephalin mutant - YVVFV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, [Leu-5]-Enkephalin mutant - YVVFV | | Authors: | Sangwan, S, Eisenberg, D, Sawaya, M.R, Do, T.D, Bowers, M.T, Lapointe, N.E, Teplow, D.B, Feinstein, S.C. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Factors that drive Peptide assembly from native to amyloid structures: experimental and theoretical analysis of [leu-5]-enkephalin mutants.
J.Phys.Chem.B, 118, 2014
|
|
3E8F
 
 | |
3VBE
 
 | |
3E7B
 
 | | Crystal Structure of Protein Phosphatase-1 Bound to the natural toxin inhibitor Tautomycin | | Descriptor: | (2Z)-2-[(1R)-3-{[(1R,2S,3R,6S,7S,10R)-10-{(2S,3S,6R,8S,9R)-3,9-dimethyl-8-[(3S)-3-methyl-4-oxopentyl]-1,7-dioxaspiro[5.5]undec-2-yl}-3,7-dihydroxy-2-methoxy-6-methyl-1-(1-methylethyl)-5-oxoundecyl]oxy}-1-hydroxy-3-oxopropyl]-3-methylbut-2-enedioic acid, AZIDE ION, CHLORIDE ION, ... | | Authors: | Kelker, M.S, Page, R, Peti, W. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: a novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors
J.Mol.Biol., 385, 2009
|
|
4OO8
 
 | | Crystal structure of Streptococcus pyogenes Cas9 in complex with guide RNA and target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*CP*CP*AP*GP*CP*CP*AP*AP*GP*CP*GP*CP*AP*CP*CP*TP*AP*AP*TP*TP*TP*CP*C)-3'), RNA (97-MER) | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2014-01-31 | | Release date: | 2014-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Cas9 in complex with guide RNA and target DNA
Cell(Cambridge,Mass.), 156, 2014
|
|
3VF2
 
 | | Crystal structure of the O-carbamoyltransferase TobZ M473I variant in complex with carbamoyl phosphate and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FE (II) ION, O-carbamoyltransferase TobZ, ... | | Authors: | Parthier, C, Stubbs, M.T, Goerlich, S, Jaenecke, F. | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3EES
 
 | |
3VPL
 
 | | Crystal structure of a 2-fluoroxylotriosyl complex of the Vibrio sp. AX-4 Beta-1,3-xylanase | | Descriptor: | 3,4-dinitrophenol, Beta-1,3-xylanase XYL4, beta-D-xylopyranose-(1-3)-beta-D-xylopyranose-(1-3)-2-deoxy-2-fluoro-beta-D-xylopyranose, ... | | Authors: | Watanabe, N, Sakaguchi, K. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of a 2-fluoroxylotriosyl complex of the Vibrio sp. AX-4 beta-1,3-xylanase at 1.2 A resolution
To be Published
|
|
4OTG
 
 | | Crystal Structure of PRK1 Catalytic Domain in Complex with Lestaurtinib | | Descriptor: | Lestaurtinib, Serine/threonine-protein kinase N1 | | Authors: | Chamberlain, P.P, Delker, S, Pagarigan, B, Mahmoudi, A, Jackson, P, Abbassian, M, Muir, J, Raheja, N, Cathers, B. | | Deposit date: | 2014-02-13 | | Release date: | 2014-08-27 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of PRK1 in Complex with the Clinical Compounds Lestaurtinib and Tofacitinib Reveal Ligand Induced Conformational Changes.
Plos One, 9, 2014
|
|
