8FE5
 
 | | Structure of J-PKAc chimera complexed with Aplithianine B | | Descriptor: | 6-[(6P)-6-(1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7,9-dihydro-8H-purin-8-one, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
1FU4
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | GLYCOGEN PHOSPHORYLASE, N-[(5S,7R,8S,9S,10R)-8,9,10-trihydroxy-7-(hydroxymethyl)-2,4-dioxo-6-oxa-1,3-diazaspiro[4.5]dec-3-yl]acetamide, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
6YOR
 
 | | Structure of the SARS-CoV-2 spike S1 protein in complex with CR3022 Fab | | Descriptor: | IgG H chain, IgG L chain, Spike glycoprotein | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Duyvesteyn, H.M.E, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-15 | | Release date: | 2020-04-29 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
8FWE
 
 | | Neck structure of Agrobacterium phage Milano, C3 symmetry | | Descriptor: | Collar sheath protein, gp13, Neck 1 protein, ... | | Authors: | Sonani, R.R, Wang, F, Esteves, N.C, Kelly, R.J, Sebastian, A, Kreutzberger, M.A.B, Leiman, P.G, Scharf, B.E, Egelman, E.H. | | Deposit date: | 2023-01-21 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Neck and capsid architecture of the robust Agrobacterium phage Milano.
Commun Biol, 6, 2023
|
|
1FTW
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | (5S,7R,8S,9S,10R)-3,8,9,10-tetrahydroxy-7-(hydroxymethyl)-6-oxa-1,3-diazaspiro[4.5]decane-2,4-dione, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-13 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
7QYD
 
 | | mosquitocidal Cry11Ba determined at pH 6.5 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Ba | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX4
 
 | | mosquitocidal Cry11Aa determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX6
 
 | | mosquitocidal Cry11Aa-E583Q determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX5
 
 | | mosquitocidal Cry11Aa-Y449F determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX7
 
 | | mosquitocidal Cry11Aa-F17Y determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7OGS
 
 | |
8RKF
 
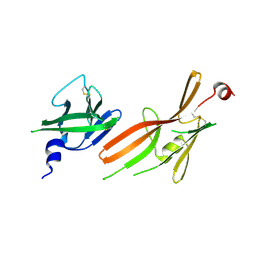 | | Crystal structure of the ZP-N1 and ZP-N2 domains of human ZP2 (hZP2-N1N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2 | | Authors: | Dioguardi, E, Stsiapanava, A, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
6V2F
 
 | | Crystal structure of the HIV capsid hexamer bound to the small molecule long-acting inhibitor, GS-6207 | | Descriptor: | HIV-1 capsid, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Appleby, T.C, Link, J.O, Yant, S.R, Villasenor, A.G, Somoza, J.R, Hu, E.Y, Schroeder, S.D, Cihlar, T. | | Deposit date: | 2019-11-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Clinical targeting of HIV capsid protein with a long-acting small molecule.
Nature, 584, 2020
|
|
4X4D
 
 | |
1FU7
 
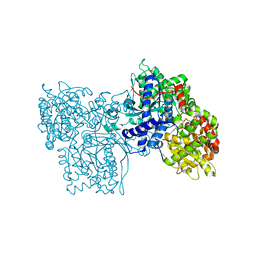 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | GLYCOGEN PHOSPHORYLASE, N-(methoxycarbonyl)-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
7OF3
 
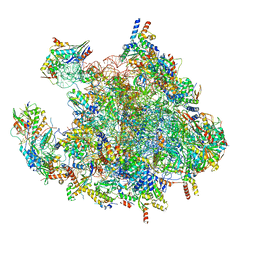 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with MTERF4-NSUN4 (dataset2). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
4X4I
 
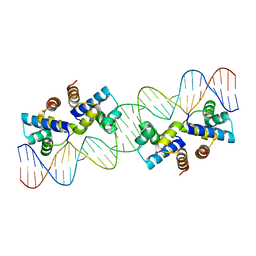 | |
4X4E
 
 | |
4X4H
 
 | |
1FU8
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 1-DEOXY-1-ACETYLAMINO-BETA-D-GLUCO-2-HEPTULOPYRANOSONAMIDE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
7OOX
 
 | | Crystal structure of PIM1 in complex with ARC-3126 | | Descriptor: | 1,2-ETHANEDIOL, Inhibitor ARC-3126, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Dixon-Clarke, S.E, Nonga, O.E, Uri, A, Bullock, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure-Guided Design of Bisubstrate Inhibitors and Photoluminescent Probes for Protein Kinases of the PIM Family.
Molecules, 26, 2021
|
|
7OOV
 
 | | Crystal structure of PIM1 in complex with ARC-1411 | | Descriptor: | 1,2-ETHANEDIOL, Inhibitor ARC-1411, SULFATE ION, ... | | Authors: | Chaikuad, A, Dixon-Clarke, S.E, Nonga, O.E, Uri, A, Bullock, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure-Guided Design of Bisubstrate Inhibitors and Photoluminescent Probes for Protein Kinases of the PIM Family.
Molecules, 26, 2021
|
|
7OOW
 
 | | Crystal structure of PIM1 in complex with ARC-1415 | | Descriptor: | 1,2-ETHANEDIOL, INHIBITOR ARC-1415, MAGNESIUM ION, ... | | Authors: | Chaikuad, A, Dixon-Clarke, S.E, Nonga, O.E, Uri, A, Bullock, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure-Guided Design of Bisubstrate Inhibitors and Photoluminescent Probes for Protein Kinases of the PIM Family.
Molecules, 26, 2021
|
|
6YWL
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, ... | | Authors: | Schroeder, M, Ni, X, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
5MDM
 
 | | Structural intermediates in the fusion associated transition of vesiculovirus glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Baquero, E, Albertini, A.A, Gaudin, Y, Bressanelli, S. | | Deposit date: | 2016-11-11 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Structural intermediates in the fusion-associated transition of vesiculovirus glycoprotein.
EMBO J., 36, 2017
|
|
