8BK5
 
 | | A structure of the truncated LpMIP with bound inhibitor JK095. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase, SODIUM ION | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8PZ8
 
 | | crystal structure of VDR in complex with D-Bishomo-1a,25-dihydroxyvitamin D3 Analog 54 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(4~{a}~{R},5~{S},9~{a}~{S})-4~{a}-methyl-5-[(2~{R})-6-methyl-6-oxidanyl-heptan-2-yl]-3,4,5,6,7,8,9,9~{a}-octahydro-2~{H}-benzo[7]annulen-1-ylidene]ethylidene]cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2023-07-27 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Design, synthesis, and biological activity of D-bishomo-1 alpha ,25-dihydroxyvitamin D 3 analogs and their crystal structures with the vitamin D nuclear receptor.
Eur.J.Med.Chem., 271, 2024
|
|
2Q1P
 
 | | Crystal Structure of Phospholipase A2 complex with propanol at 1.5 A resolution | | Descriptor: | N-PROPANOL, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Kumar, S, Hariprasad, G, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2007-05-25 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Phospholipase A2 complex with propanol at 1.5 A resolution
To be Published
|
|
8Q69
 
 | | Crystal structure of HsRNMT complexed with inhibitor DDD1060606 | | Descriptor: | 1-[(3~{S})-3-(2~{H}-pyrazolo[3,4-b]pyridin-3-yl)piperidin-1-yl]-2-thiophen-3-yl-ethanone, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-08-11 | | Release date: | 2024-08-21 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Characterisation of RNA guanine-7 methyltransferase (RNMT) using a small molecule approach.
Biochem.J., 482, 2025
|
|
4KPR
 
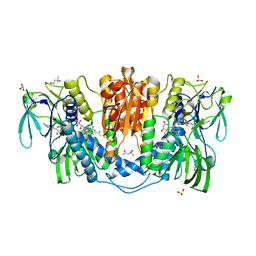 | | Tetrameric form of rat selenoprotein thioredoxin reductase 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, SULFITE ION, ... | | Authors: | Lindqvist, Y, Sandalova, T, Xu, J, Arner, E. | | Deposit date: | 2013-05-14 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Trp114 residue of thioredoxin reductase 1 is an electron relay sensor for oxidative stress
To be Published, 2013
|
|
4K3Z
 
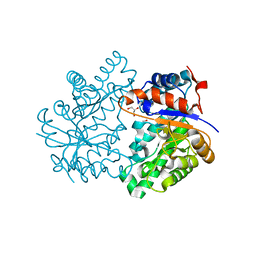 | |
8BK4
 
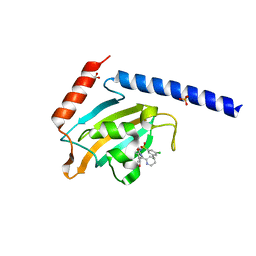 | | Full length structure of the apo-state LpMIP. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-[(1~{S})-1-pyridin-2-ylethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, GLYCEROL, Macrophage infectivity potentiator, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8BJE
 
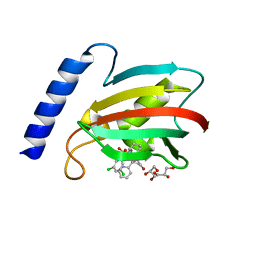 | | A structure of the truncated LpMIP with bound inhibitor JK236. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-[(1~{S})-1-pyridin-2-ylethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, GLYCEROL, Peptidyl-prolyl cis-trans isomerase | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
3AY7
 
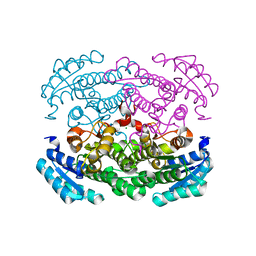 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 G259A mutant | | Descriptor: | CHLORIDE ION, Glucose 1-dehydrogenase 4 | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-04-29 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
4KFJ
 
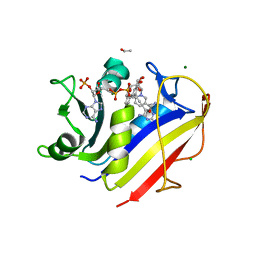 | | Human dihydrofolate reductase complexed with NADPH and 5-{3-[3-methoxy-5-(isoquin-5-yl)phenyl]prop-1-yn-1-yl}6-ethylprimidine-2,4-diamine | | Descriptor: | 6-ethyl-5-{3-[3-(isoquinolin-5-yl)-5-methoxyphenyl]prop-1-yn-1-yl}pyrimidine-2,4-diamine, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Elucidating features that drive the design of selective antifolates using crystal structures of human dihydrofolate reductase.
Biochemistry, 52, 2013
|
|
9H1M
 
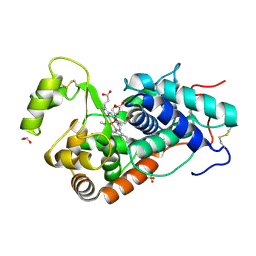 | | Recombinant ferric horseradish peroxidase C1A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Nesa, M.L, Mandal, S.K, Toelzer, C, Humer, D, Moody, P.C.E, Berger, I, Spadiut, O, Raven, E.L. | | Deposit date: | 2024-10-09 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of ferric recombinant horseradish peroxidase.
J.Biol.Inorg.Chem., 30, 2025
|
|
6CQC
 
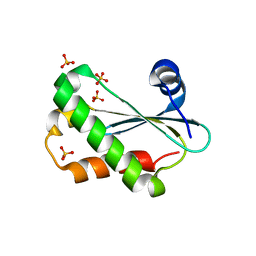 | |
4KD7
 
 | | Human dihydrofolate reductase complexed with NADPH and 5-{3-[3-methoxy-5(pyridine-4-yl)phenyl]prop-1-yn-1-yl}-6-ethyl-pyrimidine-2,4-diamine | | Descriptor: | 6-ethyl-5-{3-[3-methoxy-5-(pyridin-4-yl)phenyl]prop-1-yn-1-yl}pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2013-04-24 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.715 Å) | | Cite: | Elucidating features that drive the design of selective antifolates using crystal structures of human dihydrofolate reductase.
Biochemistry, 52, 2013
|
|
2F8A
 
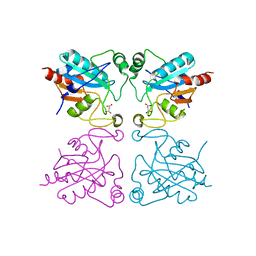 | | Crystal structure of the selenocysteine to glycine mutant of human glutathione peroxidase 1 | | Descriptor: | Glutathione peroxidase 1, MALONIC ACID | | Authors: | Kavanagh, K.L, Johansson, C, Smee, C, Gileadi, O, von Delft, F, Weigelt, J, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-02 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the selenocysteine to glycine mutant of human glutathione peroxidase 1
To be Published
|
|
4KBN
 
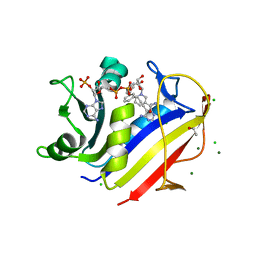 | | human dihydrofolate reductase complexed with NADPH and 5-{3-[3-(3,5-pyrimidine)]-phenyl-prop-1-yn-1-yl}-6-ethyl-pyrimidine-2,4diamine | | Descriptor: | 6-ethyl-5-{3-[3-(pyrimidin-5-yl)phenyl]prop-1-yn-1-yl}pyrimidine-2,4-diamine, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2013-04-23 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Elucidating features that drive the design of selective antifolates using crystal structures of human dihydrofolate reductase.
Biochemistry, 52, 2013
|
|
4K9X
 
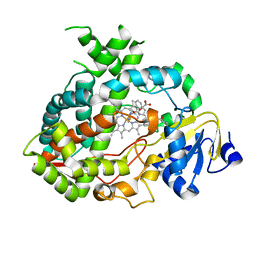 | | Complex of human CYP3A4 with a desoxyritonavir analog | | Descriptor: | 1,3-thiazol-5-ylmethyl [(2R,5R)-5-{[(2S)-2-methylbutanoyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F, Poulos, T.L. | | Deposit date: | 2013-04-21 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Dissecting Cytochrome P450 3A4-Ligand Interactions Using Ritonavir Analogues.
Biochemistry, 52, 2013
|
|
6CHU
 
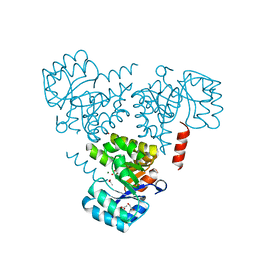 | | CRYSTAL STRUCTURE OF PROTEIN CITE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM AND ACETATE | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Citrate lyase subunit beta-like protein, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2018-02-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
9HQ6
 
 | | Structural insights in the HuHf@gold-monocarbene adduct: aurophilicity revealed in a biological context | | Descriptor: | 1-butyl-3-methyl-1H-imidazol-3-ium, Ferritin heavy chain, GOLD ION | | Authors: | Cosottini, L, Giachetti, A, Guerri, A, Martinez-Castillo, A, Geri, A, Zineddu, S, Abrescia, N.G.A, Messori, L, Turano, P, Rosato, A. | | Deposit date: | 2024-12-16 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (1.51 Å) | | Cite: | Structural Insight Into a Human H Ferritin@Gold-Monocarbene Adduct: Aurophilicity Revealed in a Biological Context.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
2EYN
 
 | | Crystal structure of the actin-binding domain of human alpha-actinin 1 at 1.8 Angstrom resolution | | Descriptor: | Alpha-actinin 1 | | Authors: | Borrego-Diaz, E, Kerff, F, Lee, S.H, Ferron, F, Li, Y, Dominguez, R. | | Deposit date: | 2005-11-09 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the actin-binding domain of alpha-actinin 1: Evaluating two competing actin-binding models.
J.Struct.Biol., 155, 2006
|
|
2EUO
 
 | | Cytochrome c peroxidase (CCP) in complex with 1-methyl-1-lambda-5-pyridin-3-yl-amine | | Descriptor: | 1-METHYL-1,6-DIHYDROPYRIDIN-3-AMINE, PROTOPORPHYRIN IX CONTAINING FE, cytochrome c peroxidase | | Authors: | Brenk, R, Vetter, S.W, Boyce, S.E, Goodin, D.B, Shoichet, B.K. | | Deposit date: | 2005-10-29 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probing molecular docking in a charged model binding site.
J.Mol.Biol., 357, 2006
|
|
2EYI
 
 | | Crystal structure of the actin-binding domain of human alpha-actinin 1 at 1.7 Angstrom resolution | | Descriptor: | Alpha-actinin 1 | | Authors: | Borrego-Diaz, E, Kerff, F, Lee, S.H, Ferron, F, Li, Y, Dominguez, R. | | Deposit date: | 2005-11-09 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the actin-binding domain of alpha-actinin 1: Evaluating two competing actin-binding models.
J.Struct.Biol., 155, 2006
|
|
2F89
 
 | | Crystal structure of human FPPS in complex with pamidronate | | Descriptor: | Farnesyl Diphosphate Synthase, MANGANESE (II) ION, PAMIDRONATE, ... | | Authors: | Rondeau, J.-M, Bitsch, F, Bourgier, E, Geiser, M, Hemmig, R, Kroemer, M, Lehmann, S, Ramage, P, Rieffel, S, Strauss, A, Green, J.R, Jahnke, W. | | Deposit date: | 2005-12-02 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the exceptional in vivo efficacy of bisphosphonate drugs.
Chemmedchem, 1, 2006
|
|
2F94
 
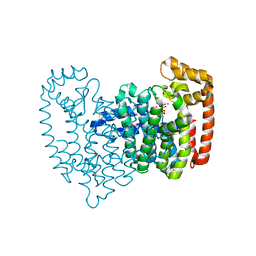 | | Crystal structure of human FPPS in complex with ibandronate | | Descriptor: | Farnesyl Diphosphate Synthase, IBANDRONATE, PHOSPHATE ION, ... | | Authors: | Rondeau, J.-M, Bitsch, F, Bourgier, E, Geiser, M, Hemmig, R, Kroemer, M, Lehmann, S, Ramage, P, Rieffel, S, Strauss, A, Green, J.R, Jahnke, W. | | Deposit date: | 2005-12-05 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for the exceptional in vivo efficacy of bisphosphonate drugs.
Chemmedchem, 1, 2006
|
|
6CHB
 
 | |
8Q0F
 
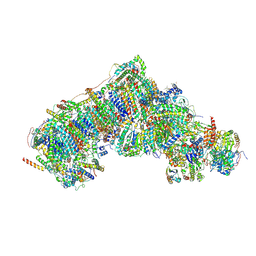 | | Inward-facing, open2 proteoliposome complex I at 3.1 A. Initially purified in DDM. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
