5J5D
 
 | |
4DPQ
 
 | | The structure of dihydrodipicolinate synthase 2 from Arabidopsis thaliana in complex with (S)-lysine | | Descriptor: | Dihydrodipicolinate synthase 2, chloroplastic, LYSINE, ... | | Authors: | Griffin, M.D.W, Billakanti, J.M, Gerrard, J.A, Dobson, R.C.J, Pearce, F.G. | | Deposit date: | 2012-02-14 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Characterisation of the first enzymes committed to lysine biosynthesis in Arabidopsis thaliana
Plos One, 7, 2012
|
|
5CZJ
 
 | |
5AFD
 
 | | Native structure of N-acetylneuramininate lyase (sialic acid aldolase) from Aliivibrio salmonicida | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-ACETYLNEURAMINATE LYASE | | Authors: | Gurung, M.K, Altermark, B, Rader, I.L.U, Helland, R, Smalas, A.O. | | Deposit date: | 2015-01-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Features and structure of a cold active N-acetylneuraminate lyase.
Plos One, 14, 2019
|
|
5KZE
 
 | | N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus | | Descriptor: | GLYCEROL, N-acetylneuraminate lyase, SULFATE ION | | Authors: | North, R.A, Watson, A.J.A, Pearce, F.G, Muscroft-Taylor, A.C, Friemann, R, Fairbanks, A.J, Dobson, R.C.J. | | Deposit date: | 2016-07-25 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure and inhibition of N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus.
FEBS Lett., 590, 2016
|
|
5KTL
 
 | |
4EOU
 
 | |
5KZD
 
 | | N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus with bound sialic acid alditol | | Descriptor: | (2~{S},4~{S},5~{R},6~{R},7~{S},8~{R})-5-acetamido-2,4,6,7,8,9-hexakis(oxidanyl)nonanoic acid, N-acetylneuraminate lyase | | Authors: | North, R.A, Watson, A.J.A, Pearce, F.G, Muscroft-Taylor, A.C, Friemann, R, Fairbanks, A.J, Dobson, R.C.J. | | Deposit date: | 2016-07-25 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Structure and inhibition of N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus.
FEBS Lett., 590, 2016
|
|
6G3Z
 
 | | Sulfolobus sulfataricus 2-keto-3-deoxygluconate (KDG) aldolase complex with D-KDPG | | Descriptor: | 2-dehydro-3-deoxy-phosphogluconate/2-dehydro-3-deoxy-6-phosphogalactonate aldolase, 2-keto 3 deoxy 6 phospho gluconate, ISOPROPYL ALCOHOL | | Authors: | Crennell, S.J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into the Substrate Specificity of Archaeal Entner-Doudoroff Aldolases: The Structures of Picrophilus torridus 2-Keto-3-deoxygluconate Aldolase and Sulfolobus solfataricus 2-Keto-3-deoxy-6-phosphogluconate Aldolase in Complex with 2-Keto-3-deoxy-6-phosphogluconate.
Biochemistry, 57, 2018
|
|
6GT8
 
 | |
6H2S
 
 | |
5C54
 
 | |
2NUY
 
 | |
2NUX
 
 | |
2OJP
 
 | | The crystal structure of a dimeric mutant of Dihydrodipicolinate synthase from E.coli- DHDPS-L197Y | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL | | Authors: | Griffin, M.D.W, Dobson, R.C.J, Antonio, L, Perugini, M.A, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2007-01-13 | | Release date: | 2008-01-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of quaternary structure in a homotetrameric enzyme.
J.Mol.Biol., 380, 2008
|
|
2NUW
 
 | |
5F1U
 
 | | biomimetic design results in a potent allosteric inhibitor of dihydrodipicolinate synthase from Campylobacter jejuni | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Conly, C.J.T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Biomimetic Design Results in a Potent Allosteric Inhibitor of Dihydrodipicolinate Synthase from Campylobacter jejuni.
J.Am.Chem.Soc., 138, 2016
|
|
7LBD
 
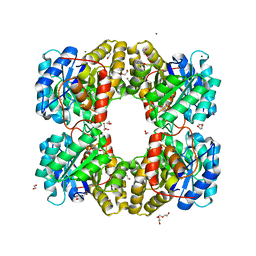 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H59K mutant with pyruvate bound in the active site in C2221 space group | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Yazdi, M.M, Sanders, D.A.R. | | Deposit date: | 2021-01-07 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Reversing the roles of a crucial hydrogen-bonding pair: a lysine-insensitive mutant of Campylobacter jejuni dihydrodipicolinate synthase, H59K, binds histidine in its allosteric site
To Be Published
|
|
7LCF
 
 | |
6UE0
 
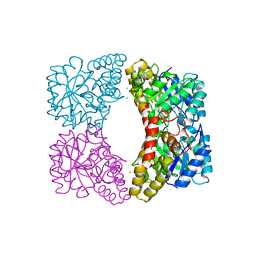 | | Crystal structure of dihydrodipicolinate synthase from Klebsiella pneumoniae bound to pyruvate | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Impey, R.E, Lee, M, Hawkins, D.A, Sutton, J.M, Panjikar, S, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Mis-annotations of a promising antibiotic target in high-priority gram-negative pathogens.
Febs Lett., 594, 2020
|
|
7M06
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, Y110F mutant with R,R-bislysine bound at the allosteric site at 2.7 Angstrom | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Saran, S, Sanders, D.A.R. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | B-FACTOR ANALYSIS SUGGEST THAT L-LYSINE AND R, R-BISLYSINE ALLOSTERICALLY INHIBIT Cj.DHDPS ENZYME BY DECREASING PROTEIN DYNAMICS
To Be Published
|
|
2A6L
 
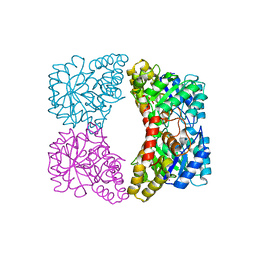 | | Dihydrodipicolinate synthase (E. coli)- mutant R138H | | Descriptor: | Dihydrodipicolinate synthase, POTASSIUM ION | | Authors: | Dobson, R.C, Devenish, S.R, Turner, L.A, Clifford, V.R, Pearce, F.G, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Role of Arginine 138 in the Catalysis and Regulation of Escherichia coli Dihydrodipicolinate Synthase.
Biochemistry, 44, 2005
|
|
6U01
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84D mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Lehnert, L, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
6TZU
 
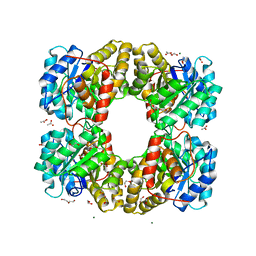 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84A mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Lehnert, C, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
2A6N
 
 | | Dihydrodipicolinate synthase (E. coli)- mutant R138A | | Descriptor: | Dihydrodipicolinate synthase, POTASSIUM ION | | Authors: | Dobson, R.C, Devenish, S.R, Turner, L.A, Clifford, V.R, Pearce, F.G, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Role of Arginine 138 in the Catalysis and Regulation of Escherichia coli Dihydrodipicolinate Synthase.
Biochemistry, 44, 2005
|
|
