6UZ5
 
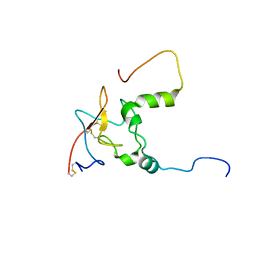 | |
6SNW
 
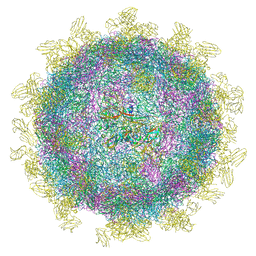 | | Structure of Coxsackievirus A10 complexed with its receptor KREMEN1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP3, ... | | Authors: | Zhao, Y, Zhou, D, Ni, T, Karia, D, Kotecha, A, Wang, X, Rao, Z, Jones, E.Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2019-08-27 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10.
Nat Commun, 11, 2020
|
|
4O03
 
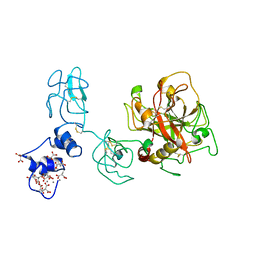 | | Crystal structure of Ca2+ bound prothrombin deletion mutant residues 146-167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Prothrombin | | Authors: | Pozzi, N, Chen, Z, Shropshire, D.B, Pelc, L.A, Di Cera, E. | | Deposit date: | 2013-12-13 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | The linker connecting the two kringles plays a key role in prothrombin activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7BZT
 
 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BZU
 
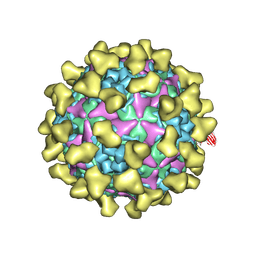 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4NZQ
 
 | | Crystal structure of Ca2+-free prothrombin deletion mutant residues 146-167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin | | Authors: | Pozzi, N, Chen, Z, Shropshire, D.B, Pelc, L.A, Di Cera, E. | | Deposit date: | 2013-12-12 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | The linker connecting the two kringles plays a key role in prothrombin activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1A0H
 
 | | THE X-RAY CRYSTAL STRUCTURE OF PPACK-MEIZOTHROMBIN DESF1: KRINGLE/THROMBIN AND CARBOHYDRATE/KRINGLE/THROMBIN INTERACTIONS AND LOCATION OF THE LINKER CHAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, MEIZOTHROMBIN | | Authors: | Martin, P.D, Malkowski, M.G, Box, J, Esmon, C.T, Edwards, B.F.P. | | Deposit date: | 1997-11-30 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | New insights into the regulation of the blood clotting cascade derived from the X-ray crystal structure of bovine meizothrombin des F1 in complex with PPACK.
Structure, 5, 1997
|
|
3E6P
 
 | | Crystal structure of human meizothrombin desF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-PHENYLALANYL-N-[(1S)-4-{[(Z)-AMINO(IMINO)METHYL]AMINO}-1-(CHLOROACETYL)BUTYL]-L-PROLINAMIDE, Prothrombin, ... | | Authors: | Papaconstantinou, M.E, Gandhi, P, Chen, Z, Bah, A, Di Cera, E. | | Deposit date: | 2008-08-15 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Na(+) binding to meizothrombin desF1.
Cell.Mol.Life Sci., 65, 2008
|
|
3K65
 
 | |
4HZH
 
 | | Structure of recombinant Gla-domainless prothrombin mutant S525A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin | | Authors: | Pozzi, N, Niu, W, Gohara, D.W, Chen, Z, Di Cera, E. | | Deposit date: | 2012-11-15 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of prothrombin reveals conformational flexibility and mechanism of activation.
J.Biol.Chem., 288, 2013
|
|
6BJR
 
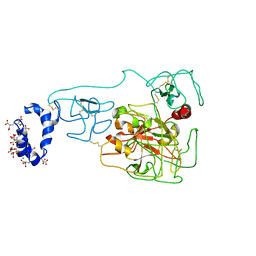 | | Crystal structure of prothrombin mutant S101C/A470C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Prothrombin, ... | | Authors: | Chinnaraj, M, Chen, Z, Pelc, L, Grese, Z, Bystranowska, D, Di Cera, E, Pozzi, N. | | Deposit date: | 2017-11-06 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structure of prothrombin in the closed form reveals new details on the mechanism of activation.
Sci Rep, 8, 2018
|
|
8UF7
 
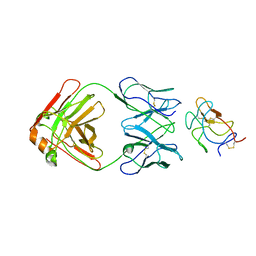 | | Cryo-EM structure of POmAb, a Type-I anti-prothrombin antiphospholipid antibody, bound to kringle-1 of human prothrombin | | Descriptor: | POmAb Heavy Chain, POmAb Light Chain, Prothrombin | | Authors: | Kumar, S, Summers, B, Basore, K, Pozzi, N. | | Deposit date: | 2023-10-03 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure and functional basis of prothrombin recognition by a type I antiprothrombin antiphospholipid antibody.
Blood, 143, 2024
|
|
6C2W
 
 | | Crystal structure of human prothrombin mutant S101C/A470C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Prothrombin, ... | | Authors: | Chinnaraj, M, Chen, Z, Pelc, L, Grese, Z, Bystranowska, D, Di Cera, E, Pozzi, N. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.12 Å) | | Cite: | Structure of prothrombin in the closed form reveals new details on the mechanism of activation.
Sci Rep, 8, 2018
|
|
2PF1
 
 | |
2PF2
 
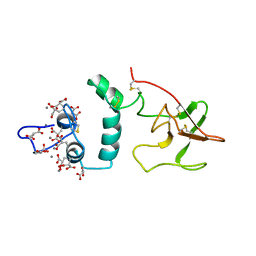 | | THE CA+2 ION AND MEMBRANE BINDING STRUCTURE OF THE GLA DOMAIN OF CA-PROTHROMBIN FRAGMENT 1 | | Descriptor: | CALCIUM ION, PROTHROMBIN FRAGMENT 1 | | Authors: | Soriano-Garcia, M, Padmanabhan, K, De vos, A.M, Tulinsky, A. | | Deposit date: | 1991-12-08 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Ca2+ ion and membrane binding structure of the Gla domain of Ca-prothrombin fragment 1.
Biochemistry, 31, 1992
|
|
5FWV
 
 | | Wnt modulator Kremen crystal form III at 3.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, KREMEN PROTEIN 1 | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
5FWS
 
 | | Wnt modulator Kremen crystal form I at 1.90A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, KREMEN PROTEIN 1, ... | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
5FWW
 
 | | Wnt modulator Kremen in complex with DKK1 (CRD2) and LRP6 (PE3PE4) | | Descriptor: | CALCIUM ION, DICKKOPF-RELATED PROTEIN 1, KREMEN PROTEIN 1, ... | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2016-09-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
5FWU
 
 | | Wnt modulator Kremen crystal form II at 2.8A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, KREMEN PROTEIN 1 | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
5FWT
 
 | | Wnt modulator Kremen crystal form I at 2.10A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, KREMEN PROTEIN 1, ... | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
2SPT
 
 | |
6OQJ
 
 | |
6OG4
 
 | | plasminogen binding group A streptococcal M protein | | Descriptor: | Plasminogen, Plasminogen-binding group A streptococcal M-like protein PAM, SULFATE ION | | Authors: | Law, R.H.P, Quek, A.J, Whisstock, J.C, Caradoc-Davies, T.T. | | Deposit date: | 2019-04-01 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Function Characterization of the a1a2 Motifs of Streptococcus pyogenes M Protein in Human Plasminogen Binding.
J.Mol.Biol., 431, 2019
|
|
6OQK
 
 | |
1NL1
 
 | | BOVINE PROTHROMBIN FRAGMENT 1 IN COMPLEX WITH CALCIUM ION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huang, M, Huang, G, Furie, B, Seaton, B, Furie, B.C. | | Deposit date: | 2003-01-06 | | Release date: | 2003-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of membrane binding by Gla domains of vitamin K-dependent proteins.
Nat.Struct.Biol., 10, 2003
|
|
