1EV6
 
 | | Structure of the monoclinic form of the M-cresol/insulin R6 hexamer | | Descriptor: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | Authors: | Smith, G.D, Ciszak, E, Magrum, L.A, Pangborn, W.A, Blessing, R.H. | | Deposit date: | 2000-04-19 | | Release date: | 2000-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | R6 Hexameric Insulin Complexed with m-Cresol or Resorcinol
Biochem.Biophys.Res.Commun., 56, 2000
|
|
6R7K
 
 | | Ligand complex of RORg LBD | | Descriptor: | (2~{R})-2-(4-ethylsulfonylphenyl)-~{N}-[4-[1,1,1,3,3,3-hexakis(fluoranyl)-2-oxidanyl-propan-2-yl]phenyl]-2-(2-phenylethanoylamino)ethanamide, DIMETHYL SULFOXIDE, Nuclear receptor ROR-gamma, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F, von Berg, S. | | Deposit date: | 2019-03-29 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Inverse Agonists of the Retinoic Acid Receptor-Related Orphan Receptor C2.
Acs Med.Chem.Lett., 10, 2019
|
|
7NPX
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 3-(3-Methoxyquinoxalin-2-yl)propanoic acid at 24 hours of soaking | | Descriptor: | 3-(3-methoxyquinoxalin-2-yl)propanoic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fata, F, Silvestri, I, Williams, D.L, Angelucci, F. | | Deposit date: | 2021-02-28 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Probing the Surface of a Parasite Drug Target Thioredoxin Glutathione Reductase Using Small Molecule Fragments.
Acs Infect Dis., 7, 2021
|
|
2GEQ
 
 | | Crystal Structure of a p53 Core Dimer Bound to DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*GP*CP*GP*TP*GP*AP*GP*CP*AP*TP*GP*CP*TP*CP*AP*C)-3', Cellular tumor antigen p53, ... | | Authors: | Ho, W.C, Fitzgerald, M.X, Marmorstein, R. | | Deposit date: | 2006-03-20 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the p53 Core Domain Dimer Bound to DNA.
J.Biol.Chem., 281, 2006
|
|
6DVS
 
 | | Crystal structure of Pseudomonas stutzeri D-phenylglycine aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Couture, J.F, Chica, R. | | Deposit date: | 2018-06-25 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Structural Determinants of the Stereoinverting Activity of Pseudomonas stutzeri d-Phenylglycine Aminotransferase.
Biochemistry, 57, 2018
|
|
8ITG
 
 | |
3VUM
 
 | | Crystal structure of a cysteine-deficient mutant M7 in MAP kinase JNK1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Mitogen-activated protein kinase 8, Peptide from C-Jun-amino-terminal kinase-interacting protein 1, ... | | Authors: | Nakaniwa, T, Kinoshita, T, Inoue, T. | | Deposit date: | 2012-07-02 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Seven cysteine-deficient mutants depict the interplay between thermal and chemical stabilities of individual cysteine residues in mitogen-activated protein kinase c-Jun N-terminal kinase 1
Biochemistry, 51, 2012
|
|
6RA6
 
 | | Ferric murine neuroglobin Gly-loop44-47/F106A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Vallone, B. | | Deposit date: | 2019-04-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lack of orientation selectivity of the heme insertion in murine neuroglobin revealed by resonance Raman spectroscopy.
Febs J., 287, 2020
|
|
5I2E
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (Hint1) with Bound Sulfamate Inhibitor 3a:3-(5-O-{[3-(1H-indol-3-yl)propanoyl]sulfamoyl}-beta-D-ribofuranosyl)-3H-imidazo[2,1-i]purine | | Descriptor: | 3-(5-O-{[3-(1H-indol-3-yl)propanoyl]sulfamoyl}-beta-D-ribofuranosyl)-3H-imidazo[2,1-i]purine, GLYCEROL, Histidine triad nucleotide-binding protein 1 | | Authors: | Strom, A.M, Finzel, B.C, Wagner, C.R. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, Synthesis, and Characterization of Sulfamide and Sulfamate Nucleotidomimetic Inhibitors of hHint1.
Acs Med.Chem.Lett., 7, 2016
|
|
8CLH
 
 | | Drug cocktail (Colchicine, Epothilone A, Peloruside, Ansamitocin P3, Vinblastine) bound to tubulin (T2R-TTL) complex | | Descriptor: | (1S,2S,3S,5S,6S,16Z,18Z,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl 2-methylpropanoate, (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, CALCIUM ION, ... | | Authors: | Wranik, M, Bertrand, Q, Kepa, M.W, Weinert, T, Steinmetz, M, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers.
Nat Commun, 14, 2023
|
|
8CLD
 
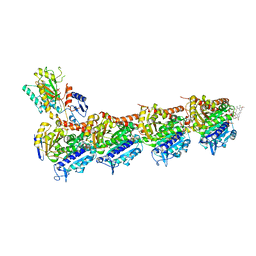 | | Ansamitocin P3 bound to tubulin (T2R-TTL) complex | | Descriptor: | (1S,2S,3S,5S,6S,16Z,18Z,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl 2-methylpropanoate, CALCIUM ION, Detyrosinated tubulin alpha-1B chain, ... | | Authors: | Wranik, M, Kepa, M.W, Bertrand, Q, Weinert, T, Steinmetz, M, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers.
Nat Commun, 14, 2023
|
|
3W0K
 
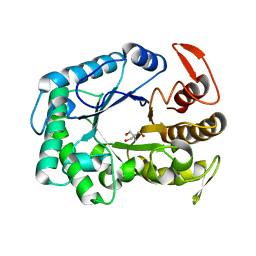 | |
8CN4
 
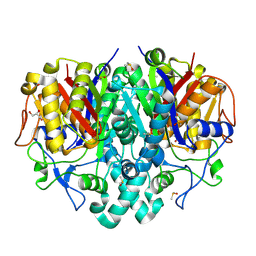 | | Pa.FabF-C164Q in complex with 5-acetamido-2-chlorobenzoic acid | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, 5-acetamido-2-chloranyl-benzoic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Georgiou, C, Brenk, R. | | Deposit date: | 2023-02-21 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | New starting points for antibiotics targeting P. aeruginosa FabF discovered by crystallographic fragment screening followed by hit expansion
Chemrxiv, 2023
|
|
6S7U
 
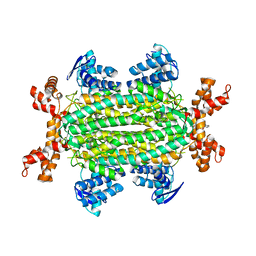 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(5-(Azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(1H-indol-3-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-06 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
6S0E
 
 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-Acetyl-2,3-dehydro-2-deoxyneuraminic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, ACETATE ION, ... | | Authors: | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | Deposit date: | 2019-06-14 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
7W7L
 
 | |
2SAK
 
 | | STAPHYLOKINASE (SAKSTAR VARIANT) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, STAPHYLOKINASE | | Authors: | Rabijns, A, De Bondt, H.L, De Maeyer, M, Lasters, I, De Ranter, C. | | Deposit date: | 1997-02-20 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of staphylokinase, a plasminogen activator with therapeutic potential.
Nat.Struct.Biol., 4, 1997
|
|
2GUP
 
 | | Structural Genomics, the crystal structure of a ROK family protein from Streptococcus pneumoniae TIGR4 in complex with sucrose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ROK family protein, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Tan, K, Li, H, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-05-01 | | Release date: | 2006-05-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The crystal structure of a ROK family protein from Streptococcus pneumoniae TIGR4 in complex with sucrose
To be Published
|
|
6LZ2
 
 | | Crystal structure of a thermostable green fluorescent protein (TGP) with a synthetic nanobody (Sb44) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Cai, H, Yao, H, Li, T, Hutter, C, Tang, Y, Li, Y, Seeger, M, Li, D. | | Deposit date: | 2020-02-17 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | An improved fluorescent tag and its nanobodies for membrane protein expression, stability assay, and purification.
Commun Biol, 3, 2020
|
|
7XGS
 
 | | Short vegetative phase protein | | Descriptor: | 1,2-ETHANEDIOL, MADS-box protein SVP | | Authors: | Liao, Y.-T, Wang, H.-C, Ko, T.-P. | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The PHYL1 effector of peanut witches broom (PnWB) phytoplasma alters the miR156/157-mediated squamosa promoter binding protein like regulation and gibberellic acid generation to promote anthocyanin biosynthesis
To Be Published
|
|
7JO2
 
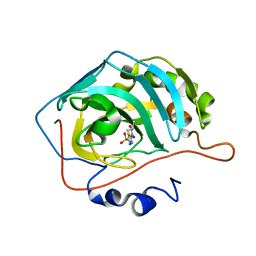 | | Carbonic Anhydrase IX Mimic Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)isobutyramide | | Descriptor: | 2-methyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.307 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
7Y0Y
 
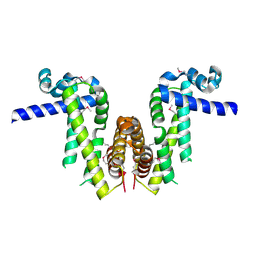 | | Crystal structure of Pseudomonas aeruginosa PvrA (SeMet) | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
5VIO
 
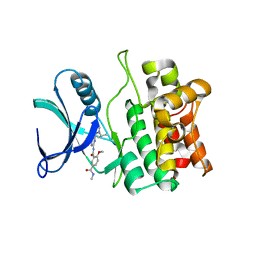 | | Crystal structure of ASK1 kinase domain with a potent inhibitor (analog 13) | | Descriptor: | 4-methoxy-N~1~-methyl-N~3~-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}benzene-1,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Jasti, J, Chang, J, Kurumbail, R. | | Deposit date: | 2017-04-17 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Rational approach to highly potent and selective apoptosis signal-regulating kinase 1 (ASK1) inhibitors.
Eur J Med Chem, 145, 2017
|
|
5VIL
 
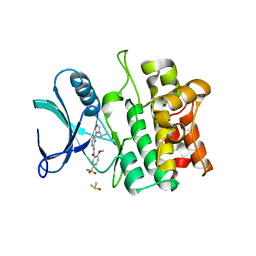 | | Crystal structure of ASK1 kinase domain with a potent inhibitor (analog 6) | | Descriptor: | 2-methoxy-N-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}-5-sulfamoylbenzamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Jasti, J, Chang, J, Kurumbail, R. | | Deposit date: | 2017-04-17 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Rational approach to highly potent and selective apoptosis signal-regulating kinase 1 (ASK1) inhibitors.
Eur J Med Chem, 145, 2017
|
|
7Y0Z
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
