4S30
 
 | |
6RW4
 
 | | Structure of human mitochondrial 28S ribosome in complex with mitochondrial IF3 | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Itoh, Y, Khawaja, A, Rorbach, J, Amunts, A. | | Deposit date: | 2019-06-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Distinct pre-initiation steps in human mitochondrial translation.
Nat Commun, 11, 2020
|
|
4GIG
 
 | |
4X3R
 
 | | Avi-GCPII structure in complex with FITC-conjugated GCPII-specific inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tykvart, J, Konvalinka, J. | | Deposit date: | 2014-12-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Design of highly potent urea-based, exosite-binding inhibitors selective for glutamate carboxypeptidase II.
J.Med.Chem., 58, 2015
|
|
4X5P
 
 | | Crystal structure of FimH in complex with a benzoyl-amidophenyl alpha-D-mannopyranoside | | Descriptor: | 4-{[3-chloro-4-(alpha-D-mannopyranosyloxy)phenyl]carbamoyl}benzoic acid, Protein FimH | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.997 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
6RMN
 
 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-05-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4X50
 
 | | Crystal structure of FimH in complex with biphenyl alpha-D-mannopyranoside | | Descriptor: | Protein FimH, biphenyl-4-yl alpha-D-mannopyranoside | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-04 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
4RLP
 
 | | Human p70s6k1 with ruthenium-based inhibitor FL772 | | Descriptor: | CHLORIDE ION, [(amino-kappaN)methanethiolato](3-fluoro-9-methoxypyrido[2,3-a]pyrrolo[3,4-c]carbazole-5,7(6H,12H)-dionato-kappa~2~N,N')(N-methyl-1,4,7-trithiecan-9-amine-kappa~3~S~1~,S~4~,S~7~)ruthenium, p70S6K1 | | Authors: | Domsic, J.F, Barber-Rotenberg, J, Salami, J, Qin, J, Marmorstein, R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Development of Organometallic S6K1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
4RM9
 
 | | Crystal structure of human ezrin in space group C2221 | | Descriptor: | Ezrin | | Authors: | Phang, J.M, Harrop, S.J, Davies, R, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2014-10-21 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin.
Biochem. J., 473, 2016
|
|
6S9W
 
 | |
4RV1
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium (NESG) Target OR497. | | Descriptor: | ACETATE ION, Engineered Protein OR497 | | Authors: | Vorobiev, S, Parmeggiani, F, Seetharaman, J, Xiao, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-11-24 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.573 Å) | | Cite: | Crystal Structure of Engineered Protein OR497.
To be Published
|
|
4X5Q
 
 | | Crystal structure of FimH in complex with 5-nitro-indolinylphenyl alpha-D-mannopyranoside | | Descriptor: | 4-(5-nitro-1H-indol-1-yl)phenyl alpha-D-mannopyranoside, Protein FimH | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
6S9X
 
 | | Crystal Structure of AKT1 in Complex with Covalent-Allosteric AKT Inhibitor 15c | | Descriptor: | RAC-alpha serine/threonine-protein kinase, ~{N}-[3-[1-[[4-[5-[(4-hydroxyphenyl)methyl]-6-oxidanylidene-2-phenyl-1~{H}-pyrazin-3-yl]phenyl]methyl]piperidin-4-yl]-2-oxidanylidene-1~{H}-benzimidazol-5-yl]propanamide | | Authors: | Landel, I, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Covalent-Allosteric Inhibitors to Achieve Akt Isoform-Selectivity.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
4G69
 
 | |
4RKW
 
 | | Crystal structure of DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Liddington, R.C. | | Deposit date: | 2014-10-14 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transnitrosylation from DJ-1 to PTEN attenuates neuronal cell death in parkinson's disease models.
J.Neurosci., 34, 2014
|
|
4RLO
 
 | | Human p70s6k1 with ruthenium-based inhibitor EM5 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Domsic, J.F, Barber-Rotenberg, J, Salami, J, Qin, J, Marmorstein, R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.527 Å) | | Cite: | Development of Organometallic S6K1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
4WUD
 
 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from no salt condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6SEH
 
 | | Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease | | Descriptor: | Structure-specific endonuclease subunit SLX1, Structure-specific endonuclease subunit SLX4, ZINC ION | | Authors: | Gaur, V, Zajko, W, Nirwal, S, Szlachcic, A, Gapinska, M, Nowotny, M. | | Deposit date: | 2019-07-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease.
Nucleic Acids Res., 47, 2019
|
|
4RU2
 
 | |
6Q7K
 
 | | ERK2 mini-fragment binding | | Descriptor: | 1H-imidazol-2-amine, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
4WX4
 
 | | Crystal structure of adenovirus 8 protease in complex with a nitrile inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCINE, N-[(2-cyanopyrimidin-4-yl)methyl]-3-[2-(3,5-dichlorophenyl)-2-methylpropanoyl]-4-methoxybenzamide, ... | | Authors: | Grosche, P, Sirockin, F, Mac Sweeney, A, Ramage, P, Erbel, P, Melkko, S, Bernardi, A, Hughes, N, Ellis, D, Combrink, K, Jarousse, N, Altmann, E. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure-based design and optimization of potent inhibitors of the adenoviral protease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6SKL
 
 | | Cryo-EM structure of the CMG Fork Protection Complex at a replication fork - Conformation 1 | | Descriptor: | Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA fork, ... | | Authors: | Yeeles, J, Baretic, D, Jenkyn-Bedford, M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork.
Mol.Cell, 78, 2020
|
|
4X22
 
 | | Crystal structure of Leptospira Interrogans Triosephosphate Isomerase (LiTIM) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Triosephosphate isomerase | | Authors: | Pareek, V, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2014-11-25 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
4WXJ
 
 | | Drosophila muscle GluRIIB complex with glutamate | | Descriptor: | GLUTAMIC ACID, Glutamate receptor IIB,Glutamate receptor IIB | | Authors: | Dharkar, P, Mayer, M.L. | | Deposit date: | 2014-11-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Functional reconstitution of Drosophila melanogaster NMJ glutamate receptors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6PTJ
 
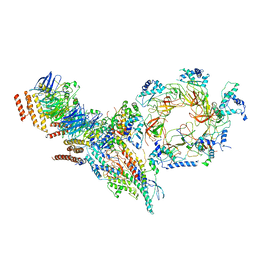 | | Structure of Ctf4 trimer in complex with one CMG helicase | | Descriptor: | Cell division control protein 45, DNA polymerase alpha-binding protein, DNA replication complex GINS protein PSF1, ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
