6ET2
 
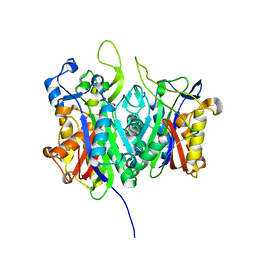 | |
1E51
 
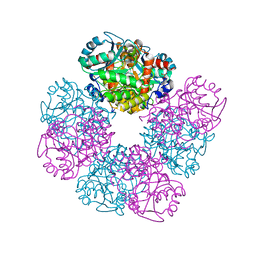 | | Crystal structure of native human erythrocyte 5-aminolaevulinic acid dehydratase | | Descriptor: | 3-[5-(AMINOMETHYL)-4-(CARBOXYMETHYL)-1H-PYRROL-3-YL]PROPANOIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, SULFATE ION, ... | | Authors: | Mills-Davies, N.L, Thompson, D, Cooper, J.B, Shoolingin-Jordan, P.M. | | Deposit date: | 2000-07-13 | | Release date: | 2001-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The Crystal Structure of Human Ala-Dehydratase
To be Published
|
|
1KLP
 
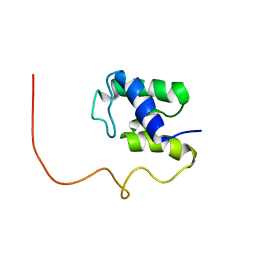 | | The Solution Structure of Acyl Carrier Protein from Mycobacterium tuberculosis | | Descriptor: | MEROMYCOLATE EXTENSION ACYL CARRIER PROTEIN | | Authors: | Wong, H.C, Liu, G, Zhang, Y.-M, Rock, C.O, Zheng, J. | | Deposit date: | 2001-12-12 | | Release date: | 2002-06-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of acyl carrier protein from Mycobacterium tuberculosis.
J.Biol.Chem., 277, 2002
|
|
7EQI
 
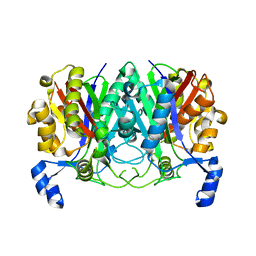 | | ChlB3 [Aceyltransferase] | | Descriptor: | ChlB3 | | Authors: | Saeed, A.U, Zheng, J. | | Deposit date: | 2021-05-02 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Insight of KSIII ( beta-Ketoacyl-ACP Synthase)-like Acyltransferase ChlB3 in the Biosynthesis of Chlorothricin.
Molecules, 27, 2022
|
|
3PXP
 
 | |
3NJ8
 
 | |
6N97
 
 | | Methylmalonyl-CoA decarboxylase in complex with 2-sulfonate-propionyl-amino(dethia)-CoA | | Descriptor: | (2R)-sulfonatepropionyl-amino(dethia)-CoA, (2S)-sulfonatepropionyl-amino(dethia)-CoA, IMIDAZOLE, ... | | Authors: | Stunkard, L.M, Dixon, A.D, Huth, T.J, Lohman, J.R. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Sulfonate/Nitro Bearing Methylmalonyl-Thioester Isosteres Applied to Methylmalonyl-CoA Decarboxylase Structure-Function Studies.
J. Am. Chem. Soc., 141, 2019
|
|
3CSI
 
 | |
3CSH
 
 | |
3CSJ
 
 | | Human glutathione s-transferase p1-1 in complex with chlorambucil | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORAMBUCIL, CHLORIDE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The anti-cancer drug chlorambucil as a substrate for the human polymorphic enzyme glutathione transferase P1-1: kinetic properties and crystallographic characterisation of allelic variants.
J.Mol.Biol., 380, 2008
|
|
4GSS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 Y108F MUTANT | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Oakley, A, Rossjohn, J, Parker, M. | | Deposit date: | 1997-01-20 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multifunctional role of Tyr 108 in the catalytic mechanism of human glutathione transferase P1-1. Crystallographic and kinetic studies on the Y108F mutant enzyme.
Biochemistry, 36, 1997
|
|
1LBK
 
 | | Crystal structure of a recombinant glutathione transferase, created by replacing the last seven residues of each subunit of the human class pi isoenzyme with the additional C-terminal helix of human class alpha isoenzyme | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, Glutathione S-transferase class pi chimaera (CODA), ... | | Authors: | Kong, G.K.W, Micaloni, C, Mazzetti, A.P, Nuccetelli, M, Antonini, G, Stella, L, McKinstry, W.J, Polekhina, G, Rossjohn, J, Federici, G, Ricci, G, Parker, M.W, Lo Bello, M. | | Deposit date: | 2002-04-04 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Engineering a new C-terminal tail in the H-site of human glutathione transferase P1-1: structural and functional consequences.
J.Mol.Biol., 325, 2003
|
|
1PGT
 
 | |
6N95
 
 | | Methylmalonyl-CoA decarboxylase in complex with 2-sulfonate-propionyl-CoA | | Descriptor: | (2R)-sulfonatepropionyl-CoA, (2S)-sulfonatepropionyl-CoA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stunkard, L.M, Dixon, A.D, Huth, T.J, Lohman, J.R. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Sulfonate/Nitro Bearing Methylmalonyl-Thioester Isosteres Applied to Methylmalonyl-CoA Decarboxylase Structure-Function Studies.
J. Am. Chem. Soc., 141, 2019
|
|
2PGT
 
 | | CRYSTAL STRUCTURE OF HUMAN GLUTATHIONE S-TRANSFERASE P1-1[V104] COMPLEXED WITH (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, ... | | Authors: | Ji, X. | | Deposit date: | 1997-02-17 | | Release date: | 1997-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the xenobiotic substrate-binding site and location of a potential non-substrate-binding site in a class pi glutathione S-transferase.
Biochemistry, 36, 1997
|
|
3HKR
 
 | | Crystal Structure of Glutathione Transferase Pi Y108V Mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2009-05-25 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influence of the H-site residue 108 on human glutathione transferase P1-1 ligand binding: structure-thermodynamic relationships and thermal stability.
Protein Sci., 18, 2009
|
|
3HJM
 
 | | Crystal structure of human Glutathione Transferase Pi Y108V mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2009-05-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Influence of the H-site residue 108 on human glutathione transferase P1-1 ligand binding: structure-thermodynamic relationships and thermal stability.
Protein Sci., 18, 2009
|
|
1NM2
 
 | | Malonyl-CoA:ACP Transacylase | | Descriptor: | ACETIC ACID, NICKEL (II) ION, malonyl CoA:acyl carrier protein malonyltransferase | | Authors: | Keatinge-Clay, A.T, Shelat, A.A, Savage, D.F, Tsai, S, Miercke, L.J.W, O'Connell III, J.D, Khosla, C, Stroud, R.M. | | Deposit date: | 2003-01-08 | | Release date: | 2003-01-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalysis, Specificity, and ACP Docking Site of
Streptomyces coelicolor Malonyl-CoA:ACP Transacylase
Structure, 11, 2003
|
|
2BDD
 
 | | Crystal Structure of Holo-ACP-synthase from Plasmodium yoelii | | Descriptor: | ACP-synthase | | Authors: | Dong, A, Melone, M, Zhao, Y, Lew, J, Koeieradzki, I, Alam, Z, Wasney, G, Vedadi, M, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-10-20 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
6POF
 
 | | Structure of human ATP citrate lyase | | Descriptor: | ATP-citrate synthase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-07-03 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5OVL
 
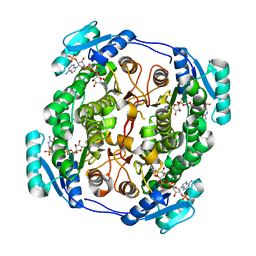 | | crystal structure of MabA bound to NADP+ from M. smegmatis | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kussau, T, Van Wyk, N, Viljoen, A, Olieric, V, Flipo, M, Kremer, L, Blaise, M. | | Deposit date: | 2017-08-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis beta-ketoacyl-acyl carrier protein reductase MabA.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6G2H
 
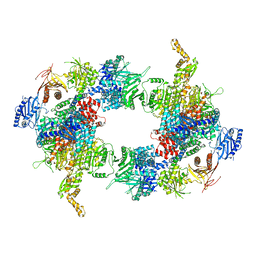 | | Filament of acetyl-CoA carboxylase and BRCT domains of BRCA1 (ACC-BRCT) core at 4.6 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1 | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
6P9L
 
 | |
6P9K
 
 | |
4DFE
 
 | |
