2IRM
 
 | | Crystal structure of mitogen-activated protein kinase kinase kinase 7 interacting protein 1 from Anopheles gambiae | | Descriptor: | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | | Authors: | Jin, X, Bonanno, J.B, Pelletier, L, Freeman, J.C, Wasserman, S, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-15 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
3WPB
 
 | | Crystal structure of horse TLR9 (unliganded form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Ohto, U, Tanji, H, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3WFS
 
 | | tRNA processing enzyme complex 3 | | Descriptor: | Poly A polymerase, RNA (74-MER), SULFATE ION | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.311 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
4AQD
 
 | | Crystal structure of fully glycosylated human butyrylcholinesterase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Wandhammer, M, Ronco, C, Trovaslet, M, Jean, L, Lockridge, O, Renard, P.Y, Nachon, F. | | Deposit date: | 2012-04-16 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human butyrylcholinesterase produced in insect cells: huprine-based affinity purification and crystal structure.
FEBS J., 279, 2012
|
|
4ADG
 
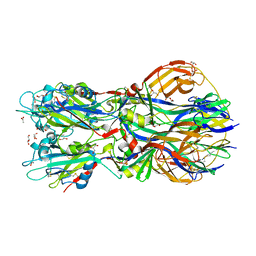 | | Crystal structure of the Rubella virus envelope Glycoprotein E1 in post-fusion form (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | DuBois, R.M, Vaney, M.C, Tortorici, M.A, Al Kurdi, R, Barba-Spaeth, G, Rey, F.A. | | Deposit date: | 2011-12-26 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Functional and Evolutionary Insight from the Crystal Structure of Rubella Virus Protein E1.
Nature, 493, 2013
|
|
4A1S
 
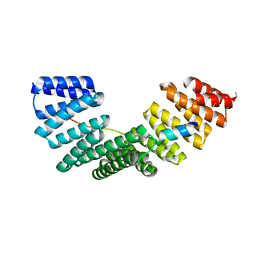 | | Crystallographic structure of the Pins:Insc complex | | Descriptor: | PARTNER OF INSCUTEABLE, RE60102P | | Authors: | Culurgioni, S, Alfieri, A, Pendolino, V, Laddomada, F, Mapelli, M. | | Deposit date: | 2011-09-19 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inscuteable and Numa Proteins Bind Competitively to Leu-Gly- Asn Repeat-Enriched Protein (Lgn) During Asymmetric Cell Divisions.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4ADI
 
 | | Crystal structure of the Rubella virus envelope glycoprotein E1 in post-fusion form (crystal form I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | DuBois, R.M, Vaney, M.C, Tortorici, M.A, Al Kurdi, R, Barba-Spaeth, G, Rey, F.A. | | Deposit date: | 2011-12-26 | | Release date: | 2013-01-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and Evolutionary Insight from the Crystal Structure of Rubella Virus Protein E1.
Nature, 493, 2013
|
|
4ANJ
 
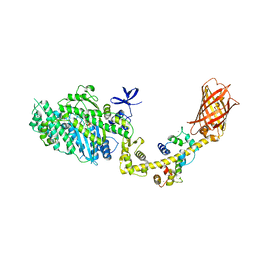 | | MYOSIN VI (MDinsert2-GFP fusion) PRE-POWERSTROKE STATE (MG.ADP.AlF4) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CALMODULIN, ... | | Authors: | Menetrey, J, Isabet, T, Ropars, V, Mukherjea, M, Pylypenko, O, Liu, X, Perez, J, Vachette, P, Sweeney, H.L, Houdusse, A.M. | | Deposit date: | 2012-03-19 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Processive Steps in the Reverse Direction Require Uncoupling of the Lead Head Lever Arm of Myosin Vi.
Mol.Cell, 48, 2012
|
|
4ADJ
 
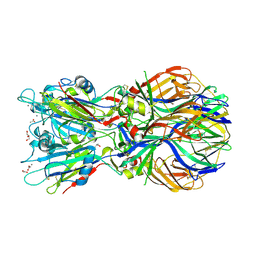 | | Crystal structure of the Rubella virus glycoprotein E1 in its post-fusion form crystallized in presence of 1mM of calcium acetate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | DuBois, R.M, Vaney, M.C, Tortorici, M.A, Al Kurdi, R, Barba-Spaeth, G, Rey, F.A. | | Deposit date: | 2011-12-26 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Functional and Evolutionary Insight from the Crystal Structure of Rubella Virus Protein E1.
Nature, 493, 2013
|
|
3C9C
 
 | | Structural Basis of Histone H4 Recognition by p55 | | Descriptor: | CADMIUM ION, Chromatin assembly factor 1 p55 subunit, Histone H4, ... | | Authors: | Song, J.J, Garlick, J.D, Kingston, R.E. | | Deposit date: | 2008-02-15 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of histone H4 recognition by p55.
Genes Dev., 22, 2008
|
|
2I0O
 
 | | Crystal structure of Anopheles gambiae Ser/Thr phosphatase complexed with Zn2+ | | Descriptor: | Ser/Thr phosphatase, ZINC ION | | Authors: | Jin, X, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
3WFR
 
 | | tRNA processing enzyme complex 2 | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
3WPF
 
 | | Crystal structure of mouse TLR9 (unliganded form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 9 | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3WPG
 
 | |
3W7B
 
 | | Crystal structure of formyltetrahydrofolate deformylase from Thermus thermophilus HB8 | | Descriptor: | Formyltetrahydrofolate deformylase | | Authors: | Sampei, G, Yanagida, Y, Ogata, N, Kusano, M, Terao, K, Kawai, H, Fukai, Y, Kanagawa, M, Inoue, Y, Baba, S, Kawai, G. | | Deposit date: | 2013-02-28 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures and reaction mechanisms of the two related enzymes, PurN and PurU
J.Biochem., 154, 2013
|
|
4ARR
 
 | | Crystal structure of the N-terminal domain of Drosophila Toll receptor with the magic triangle I3C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, TOLL RECEPTOR, ... | | Authors: | Gangloff, M, Gay, N.J. | | Deposit date: | 2012-04-26 | | Release date: | 2013-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Liesegang-Like Patterns of Toll Crystals Grown in Gel.
J.Appl.Crystallogr., 46, 2013
|
|
3WPI
 
 | | Crystal structure of mouse TLR9 in complex with inhibitory DNA_super | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*CP*CP*TP*CP*AP*AP*TP*AP*GP*GP*GP*TP*GP*AP*GP*GP*GP*G)-3'), Toll-like receptor 9 | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3WPE
 
 | |
4B3V
 
 | | Crystal structure of the Rubella virus glycoprotein E1 in its post-fusion form crystallized in presence of 20mM of Calcium Acetate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Vaney, M.C, DuBois, R.M, Tortorici, M.A, Rey, F.A. | | Deposit date: | 2012-07-26 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Functional and Evolutionary Insight from the Crystal Structure of Rubella Virus Protein E1.
Nature, 493, 2013
|
|
3WPH
 
 | |
2K56
 
 | |
4BF7
 
 | | Emericilla nidulans endo-beta-1,4-galactanase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ARABINOGALACTAN ENDO-1,4-BETA-GALACTOSIDASE A, ... | | Authors: | Otten, H, Michalak, M, Larsen, S, Mikkelsen, J.D. | | Deposit date: | 2013-03-15 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The Binding of Zinc Ions to Emericella Nidulans Endo-[Beta]-1,4-Galactanase is Essential for Crystal Formation
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1YPP
 
 | | ACID ANHYDRIDE HYDROLASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, MANGANESE (II) ION, PHOSPHATE ION | | Authors: | Harutyunyan, E.H, Kuranova, I.P, Lamzin, V.S, Dauter, Z, Wilson, K.S. | | Deposit date: | 1996-05-29 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of yeast inorganic pyrophosphatase complexed with manganese and phosphate.
Eur.J.Biochem., 239, 1996
|
|
2ERB
 
 | | AgamOBP1, and odorant binding protein from Anopheles gambiae complexed with PEG | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, MAGNESIUM ION, odorant binding protein | | Authors: | Wogulis, M, Morgan, T, Ishida, Y, Leal, W.S, Wilson, D.K. | | Deposit date: | 2005-10-24 | | Release date: | 2005-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of an odorant binding protein from Anopheles gambiae: Evidence for a common ligand release mechanism.
Biochem.Biophys.Res.Commun., 339, 2006
|
|
2CH2
 
 | | Structure of the Anopheles gambiae 3-hydroxykynurenine transaminase in complex with inhibitor | | Descriptor: | 3-HYDROXYKYNURENINE TRANSAMINASE, 4-(2-AMINOPHENYL)-4-OXOBUTANOIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rossi, F, Garavaglia, S, Giovenzana, G.B, Arca, B, Li, J, Rizzi, M. | | Deposit date: | 2006-03-10 | | Release date: | 2006-03-28 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Anopheles Gambiae 3-Hydroxykynurenine Transaminase
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
