4YIV
 
 | | Crystal structure of engineered TgAMA1 lacking the DII loop | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Apical membrane antigen AMA1, CADMIUM ION, ... | | Authors: | Parker, M.L, Boulanger, M.J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | An Extended Surface Loop on Toxoplasma gondii Apical Membrane Antigen 1 (AMA1) Governs Ligand Binding Selectivity.
Plos One, 10, 2015
|
|
3QX5
 
 | | Thrombin Inhibition by Pyridin Derivatives | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(4-chloro-1-methylpyridinium-2-yl)methyl]-L-prolinamide, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-03-01 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin
J.Mol.Biol., 418, 2012
|
|
1UY0
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with glc-1,3-glc-1,4-glc-1,3-glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
3BKR
 
 | |
4PUU
 
 | |
2F47
 
 | | Xray crystal structure of T4 lysozyme mutant L20/R63A liganded to methylguanidinium | | Descriptor: | 1-METHYLGUANIDINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Yousef, M.S, Bischoff, N, Dyer, C.M, Baase, W.A, Matthews, B.W. | | Deposit date: | 2005-11-22 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanidinium derivatives bind preferentially and trigger long-distance conformational changes in an engineered T4 lysozyme.
Protein Sci., 15, 2006
|
|
1UZ0
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with Glc-4Glc-3Glc-4Glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
4H2W
 
 | | Crystal structure of engineered Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with carrier protein from Agrobacterium tumefaciens and AMP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE MONOPHOSPHATE, Amino acid--[acyl-carrier-protein] ligase 1, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
4QBX
 
 | |
2NYJ
 
 | | Crystal structure of the ankyrin repeat domain of TRPV1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Jin, X, Gaudet, R. | | Deposit date: | 2006-11-20 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Ankyrin Repeats of TRPV1 Bind Multiple Ligands and Modulate Channel Sensitivity.
Neuron, 54, 2007
|
|
1UYZ
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with xylotetraose | | Descriptor: | CALCIUM ION, CELLULASE B, GLYCEROL, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UYX
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with cellobiose | | Descriptor: | CALCIUM ION, CELLULASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
5ZWF
 
 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having a enone with a beta methyl group via conjugate addition reaction | | Descriptor: | (E,7R)-7-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-2-en-4-one, 13-meric peptide from DRIP205 NR2 BOX peptide, Vitamin D3 receptor | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
3II7
 
 | | Crystal structure of the kelch domain of human KLHL7 | | Descriptor: | 1,2-ETHANEDIOL, Kelch-like protein 7 | | Authors: | Chaikuad, A, Thangaratnarajah, C, Cooper, C.D.O, Ugochukwu, E, Muniz, J.R.C, Krojer, T, Sethi, R, Pike, A.C.W, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases.
J.Biol.Chem., 288, 2013
|
|
2V8P
 
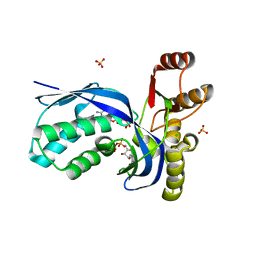 | | IspE in complex with ADP and CDP | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL KINASE, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Sgraja, T, Alphey, M.S, Hunter, W.N. | | Deposit date: | 2007-08-13 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of Aquifex Aeolicus 4-Diphosphocytidyl-2C-Methyl-D-Erythritol Kinase -Ligand Recognition in a Template for Antimicrobial Drug Discovery.
FEBS J., 275, 2008
|
|
2ZGB
 
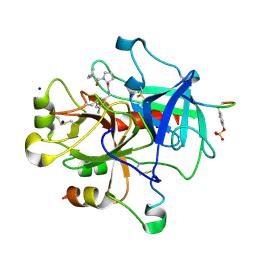 | | Thrombin Inhibition | | Descriptor: | D-leucyl-N-(3-chlorobenzyl)-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-01-21 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Non-additivity of functional group contributions in protein-ligand binding: a comprehensive study by crystallography and isothermal titration calorimetry.
J.Mol.Biol., 397, 2010
|
|
2ZI2
 
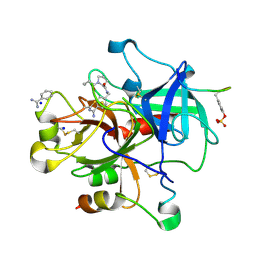 | | Thrombin Inhibition | | Descriptor: | 1-butanoyl-N-(4-carbamimidoylbenzyl)-L-prolinamide, BENZAMIDINE, Hirudin variant-1, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-02-13 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Non-additivity of functional group contributions in protein-ligand binding: a comprehensive study by crystallography and isothermal titration calorimetry.
J.Mol.Biol., 397, 2010
|
|
4Q9H
 
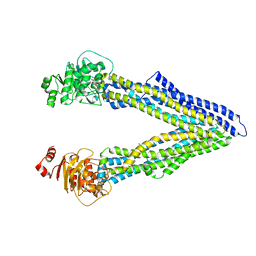 | | P-glycoprotein at 3.4 A resolution | | Descriptor: | Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1K21
 
 | | HUMAN THROMBIN-INHIBITOR COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin variant-2, Prothrombin, ... | | Authors: | Stubbs, M.T, Musil, D. | | Deposit date: | 2001-09-26 | | Release date: | 2002-05-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
2HNV
 
 | | Crystal Structure of a Dipeptide Complex of the Q58V Mutant of Bovine Neurophysin-I | | Descriptor: | Oxytocin-neurophysin 1, PHENYLALANINE, TYROSINE | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
8XD6
 
 | | Cryo-EM structure of Glutamate dehydrogenase from Thermococcus profundus in complex with NADPH, AKG and NH4 in the steady stage of reaction | | Descriptor: | 2-OXOGLUTARIC ACID, AMMONIUM ION, Glutamate dehydrogenase, ... | | Authors: | Wakabayashi, T, Oide, M, Nakasako, M. | | Deposit date: | 2023-12-10 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | CryoEM-sampling of metastable conformations appearing in cofactor-ligand association and catalysis of glutamate dehydrogenase.
Sci Rep, 14, 2024
|
|
9B91
 
 | |
9B90
 
 | |
9EQH
 
 | | WWP2 WW2-2,3-linker-HECT (WWP2-LH) | | Descriptor: | GLYCEROL, Isoform 2 of NEDD4-like E3 ubiquitin-protein ligase WWP2, SODIUM ION | | Authors: | Dudey, A.P, Hemmings, A.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Expanding the Inhibitor Space of the WWP1 and WWP2 HECT E3 Ligases
To Be Published
|
|
6MU1
 
 | | Structure of full-length IP3R1 channel bound with Adenophostin A | | Descriptor: | Adenophostin A, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Serysheva, I.I, Fan, G, Baker, M.R, Wang, Z, Seryshev, A, Ludtke, S.J, Baker, M.L. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM reveals ligand induced allostery underlying InsP3R channel gating.
Cell Res., 28, 2018
|
|
