5BQD
 
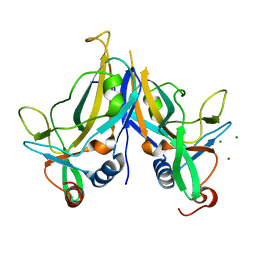 | | Crystal Structure of TBX5 (1-239) Dimer | | Descriptor: | MAGNESIUM ION, T-box transcription factor TBX5 | | Authors: | Pradhan, L, Gopal, S, Patel, A, Kasahara, H, Nam, H.J. | | Deposit date: | 2015-05-28 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | Intermolecular Interactions of Cardiac Transcription Factors NKX2.5 and TBX5.
Biochemistry, 55, 2016
|
|
4MVJ
 
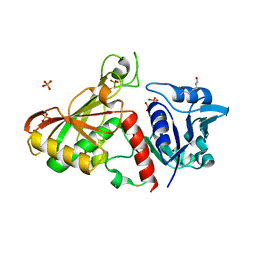 | | 2.85 Angstrom Resolution Crystal Structure of Glyceraldehyde 3-phosphate Dehydrogenase A (gapA) from Escherichia coli Modified by Acetyl Phosphate. | | Descriptor: | ACETATE ION, ACETYLPHOSPHATE, CHLORIDE ION, ... | | Authors: | Minasov, G, Kuhn, M, Dubrovska, I, Winsor, J, Shuvalova, L, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
Plos One, 9, 2014
|
|
1ZBA
 
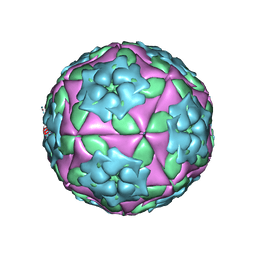 | | Foot-and-Mouth Disease virus serotype A1061 complexed with oligosaccharide receptor. | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Coat protein VP1, Coat protein VP2, ... | | Authors: | Fry, E.E, Newman, J.W, Curry, S, Najjam, S, Jackson, T, Blakemore, W, Lea, S.M, Miller, L, Burman, A, King, A.M, Stuart, D.I. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Foot-and-mouth disease virus serotype A1061 alone and complexed with oligosaccharide receptor: receptor conservation in the face of antigenic variation.
J.Gen.Virol., 86, 2005
|
|
1ZBE
 
 | | Foot-and Mouth Disease Virus Serotype A1061 | | Descriptor: | Coat protein VP1, Coat protein VP2, Coat protein VP3, ... | | Authors: | Fry, E.E, Newman, J.W, Curry, S, Najjam, S, Jackson, T, Blakemore, W, Lea, S.M, Miller, L, Burman, A, King, A.M, Stuart, D.I. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Foot-and-mouth disease virus serotype A1061 alone and complexed with oligosaccharide receptor: receptor conservation in the face of antigenic variation.
J.Gen.Virol., 86, 2005
|
|
3IIM
 
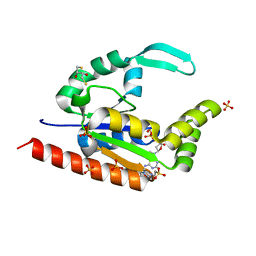 | | The structure of hCINAP-dADP complex at 2.0 angstroms resolution | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Zographos, S.E, Drakou, C.E, Leonidas, D.D. | | Deposit date: | 2009-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | hCINAP is an atypical mammalian nuclear adenylate kinase with an ATPase motif: Structural and functional studies.
Proteins, 80, 2012
|
|
3QB7
 
 | | Interleukin-4 mutant RGA bound to cytokine receptor common gamma | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, ... | | Authors: | Bates, D.L, Junttila, I.S, Creusot, R.J, Moraga, I, Lupardus, P, Fathman, C.G, Paul, W.E, Garcia, K.C. | | Deposit date: | 2011-01-12 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.245 Å) | | Cite: | Redirecting cell-type specific cytokine responses with engineered interleukin-4 superkines.
Nat.Chem.Biol., 8, 2012
|
|
2XYA
 
 | | Non-covalent inhibtors of rhinovirus 3C protease. | | Descriptor: | 2-PHENYLQUINOLIN-4-OL, PICORNAIN 3C | | Authors: | Petersen, J, Edman, K, Edfeldt, F, Johansson, C. | | Deposit date: | 2010-11-16 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Non-Covalent Inhibitors of Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3IIJ
 
 | |
3IIK
 
 | |
6VKQ
 
 | | Crystal Structure of human PARP-1 CAT domain bound to inhibitor EB-47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Steffen, J.D, Pascal, J.M. | | Deposit date: | 2020-01-21 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for allosteric PARP-1 retention on DNA breaks.
Science, 368, 2020
|
|
6VKK
 
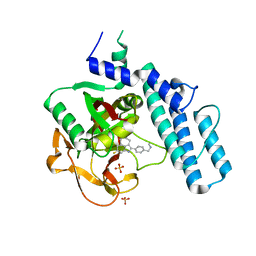 | |
1PIV
 
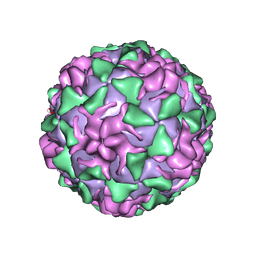 | | BINDING OF THE ANTIVIRAL DRUG WIN51711 TO THE SABIN STRAIN OF TYPE 3 POLIOVIRUS: STRUCTURAL COMPARISON WITH DRUG BINDING IN RHINOVIRUS 14 | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, POLIOVIRUS TYPE 3 (SUBUNIT VP1), ... | | Authors: | Hiremath, C.N, Grant, R.A, Filman, D.J, Hogle, J.M. | | Deposit date: | 1995-02-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Binding of the antiviral drug WIN51711 to the sabin strain of type 3 poliovirus: structural comparison with drug binding in rhinovirus 14.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
3IIL
 
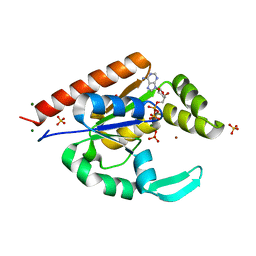 | | The structure of hCINAP-MgADP-Pi complex at 2.0 angstroms resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Coilin-interacting nuclear ATPase protein, LITHIUM ION, ... | | Authors: | Zographos, S.E, Drakou, C.E, Leonidas, D.D. | | Deposit date: | 2009-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | hCINAP is an atypical mammalian nuclear adenylate kinase with an ATPase motif: Structural and functional studies.
Proteins, 80, 2012
|
|
6VKO
 
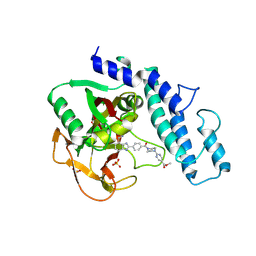 | |
3RI4
 
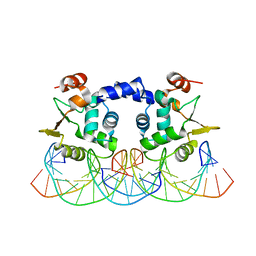 | |
1XO1
 
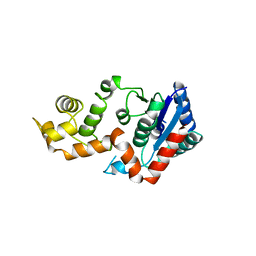 | | T5 5'-EXONUCLEASE MUTANT K83A | | Descriptor: | 5'-EXONUCLEASE | | Authors: | Ceska, T.A, Suck, D, Sayers, J.R. | | Deposit date: | 1998-11-19 | | Release date: | 1999-04-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis of conserved lysine residues in bacteriophage T5 5'-3' exonuclease suggests separate mechanisms of endo-and exonucleolytic cleavage.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3QB1
 
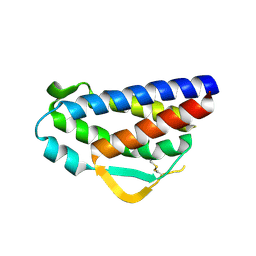 | | Interleukin-2 mutant D10 | | Descriptor: | Interleukin-2 | | Authors: | Levin, A.M, Bates, D.L, Ring, A.M, Lin, J.T, Su, L, Krieg, C, Bowman, G.R, Novick, P, Pande, V.S, Khort, H.E, Boyman, O, Fathman, C.G, Garcia, K.C. | | Deposit date: | 2011-01-12 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Exploiting a natural conformational switch to engineer an interleukin-2 'superkine'
Nature, 484, 2012
|
|
3WE1
 
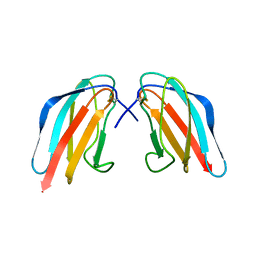 | | Crystal structure of Dengue 4 Envelope protein domain III (ED3) | | Descriptor: | Envelope protein E | | Authors: | Elahi, M, Islam, M.M, Noguchi, K, Yohda, M, Toh, H, Kuroda, Y. | | Deposit date: | 2013-06-27 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.278 Å) | | Cite: | Computational prediction and experimental characterization of a "size switch type repacking" during the evolution of dengue envelope protein domain III (ED3).
Biochim.Biophys.Acta, 1844, 2014
|
|
2MXD
 
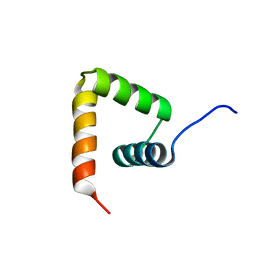 | | Solution structure of VPg of porcine sapovirus | | Descriptor: | Viral protein genome-linked | | Authors: | Kim, J, Hwang, H, Min, H, Yun, H, Cho, K, Pelton, J.G, Wemmer, D.E, Lee, C. | | Deposit date: | 2014-12-24 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the porcine sapovirus VPg core reveals a stable three-helical bundle with a conserved surface patch.
Biochem.Biophys.Res.Commun., 459, 2015
|
|
1SFK
 
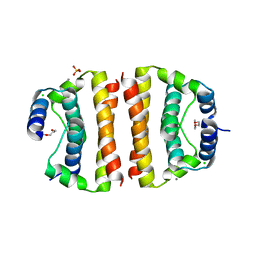 | | Core (C) protein from West Nile Virus, subtype Kunjin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Core protein, ... | | Authors: | Dokland, T, Walsh, M, Mackenzie, J.M, Khromykh, A.A, Ee, K.-H, Wang, S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | West nile virus core protein; tetramer structure and ribbon formation
Structure, 12, 2004
|
|
2HU2
 
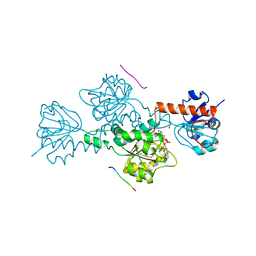 | | CTBP/BARS in ternary complex with NAD(H) and RRTGAPPAL peptide | | Descriptor: | 9-mer peptide from Zinc finger protein 217, C-terminal-binding protein 1, FORMIC ACID, ... | | Authors: | Nardini, M, Bolognesi, M, Quinlan, K.G.R, Verger, A, Francescato, P, Crossley, M. | | Deposit date: | 2006-07-26 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Specific Recognition of ZNF217 and Other Zinc Finger Proteins at a Surface Groove of C-Terminal Binding Proteins
Mol.Cell.Biol., 26, 2006
|
|
3JVK
 
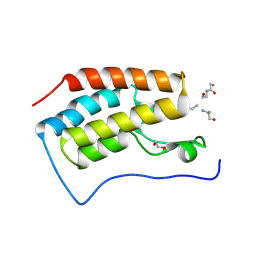 | | Crystal structure of bromodomain 1 of mouse Brd4 in complex with histone H3-K(ac)14 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, histone H3.3 peptide | | Authors: | Vollmuth, F, Blankenfeldt, W, Geyer, M. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Dual Bromodomains of the P-TEFb-activating Protein Brd4 at Atomic Resolution
J.Biol.Chem., 284, 2009
|
|
1KXA
 
 | |
1F9L
 
 | |
1D4M
 
 | | THE CRYSTAL STRUCTURE OF COXSACKIEVIRUS A9 TO 2.9 A RESOLUTION | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, PROTEIN (COXSACKIEVIRUS A9) | | Authors: | Hendry, E, Hatanaka, H, Fry, E, Smyth, M, Tate, J, Stanway, G, Santti, J, Maaronen, M, Hyypia, T, Stuart, D. | | Deposit date: | 1999-10-04 | | Release date: | 1999-12-23 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses.
Structure Fold.Des., 7, 1999
|
|
