2XRW
 
 | | Linear binding motifs for JNK and for calcineurin antagonistically control the nuclear shuttling of NFAT4 | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE 8, NUCLEAR FACTOR OF ACTIVATED T-CELLS, ... | | Authors: | Barkai, T, Toeoroe, I, Garai, A, Remenyi, A. | | Deposit date: | 2010-09-23 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Specificity of Linear Motifs that Bind to a Common Mitogen-Activated Protein Kinase Docking Groove.
Sci. Signal, 5, 2012
|
|
2XCQ
 
 | | The 2.98A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase | | Descriptor: | DNA GYRASE SUBUNIT B, DNA GYRASE SUBUNIT A | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-24 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
8RTY
 
 | | Structure of the F-actin barbed end bound by Cdc12 and profilin (ring complex) at a resolution of 6.3 Angstrom | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
2XT3
 
 | | HUMAN KIF7, A KINESIN INVOLVED IN HEDGEHOG SIGNALLING | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KIF7, MAGNESIUM ION | | Authors: | Klejnot, M, Kozielski, F. | | Deposit date: | 2010-10-04 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Structural Insights Into Human Kif7, a Kinesin Involved in Hedgehog Signalling.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2FUT
 
 | | Crystal Structure of Heparinase II Complexed with a Disaccharide Product | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ZINC ION, heparinase II protein | | Authors: | Shaya, D, Cygler, M. | | Deposit date: | 2006-01-27 | | Release date: | 2006-04-18 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Heparinase II from Pedobacter heparinus and Its Complex with a Disaccharide Product.
J.Biol.Chem., 281, 2006
|
|
4FFP
 
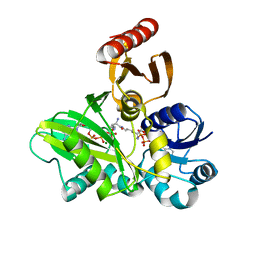 | | PylC in complex with L-lysine-Ne-D-ornithine (cocrystallized with L-lysine and D-ornithine) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, N~6~-D-ornithyl-L-lysine, ... | | Authors: | Quitterer, F, List, A, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of PylC at 1.5A resolution.
J.Mol.Biol., 424, 2012
|
|
2XH0
 
 | |
8S3E
 
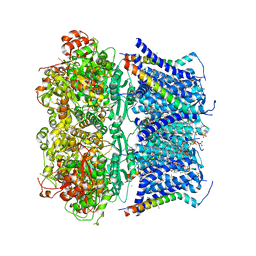 | | Structure of rabbit Slo1 in complex with gamma1/LRRC26 | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Redhardt, M, Raunser, S, Raisch, T. | | Deposit date: | 2024-02-20 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Cryo-EM structure of the Slo1 potassium channel with the auxiliary gamma 1 subunit suggests a mechanism for depolarization-independent activation.
Febs Lett., 598, 2024
|
|
2XHZ
 
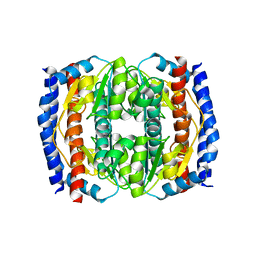 | | Probing the active site of the sugar isomerase domain from E. coli arabinose-5-phosphate isomerase via X-ray crystallography | | Descriptor: | ARABINOSE 5-PHOSPHATE ISOMERASE | | Authors: | Gourlay, L.J, Sommaruga, S, Nardini, M, Sperandeo, P, Deho, G, Polissi, A, Bolognesi, M. | | Deposit date: | 2010-06-24 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Probing the Active Site of the Sugar Isomerase Domain from E. Coli Arabinose-5-Phosphate Isomerase Via X-Ray Crystallography.
Protein Sci., 19, 2010
|
|
2XIR
 
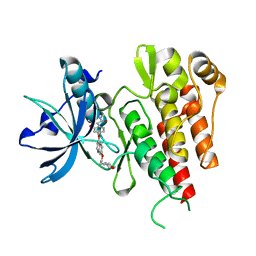 | | Crystal structure of the VEGFR2 kinase domain in complex with PF- 00337210 (N,2-dimethyl-6-(7-(2-morpholinoethoxy)quinolin-4-yloxy) benzofuran-3-carboxamide) | | Descriptor: | N,2-DIMETHYL-6-{[7-(2-MORPHOLIN-4-YLETHOXY)QUINOLIN-4-YL]OXY}-1-BENZOFURAN-3-CARBOXAMIDE, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 2 | | Authors: | McTigue, M, Wickersham, J, Pinko, C, Hong, Y, Marrone, T. | | Deposit date: | 2010-06-30 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of the Selective Vegfr Inhibitor Pf- 00337210
To be Published
|
|
2XYV
 
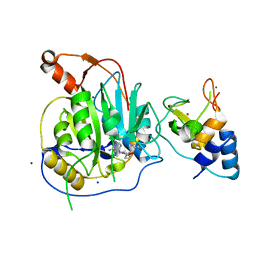 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-19 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2XJ8
 
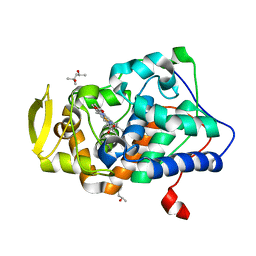 | | The structure of ferrous cytochrome c peroxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-07-02 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
2XM5
 
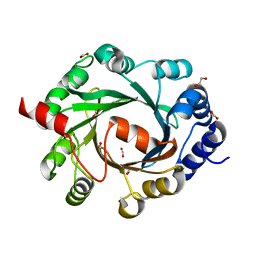 | | Structural and Mechanistic Analysis of the Magnesium-Independent Aromatic Prenyltransferase CloQ from the Clorobiocin Biosynthetic Pathway | | Descriptor: | 1,2-ETHANEDIOL, CLOQ, FORMIC ACID | | Authors: | Metzger, U, Keller, S, Stevenson, C.E.M, Heide, L, Lawson, D.M. | | Deposit date: | 2010-07-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Mechanism of the Magnesium-Independent Aromatic Prenyltransferase Cloq from the Clorobiocin Biosynthetic Pathway.
J.Mol.Biol., 404, 2010
|
|
8R5O
 
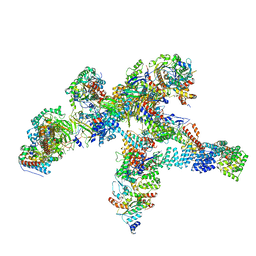 | | Plastid-encoded RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
2XNQ
 
 | |
8RC1
 
 | |
2Y0H
 
 | |
2XBR
 
 | | Raman crystallography of Hen White Egg Lysozyme - Low X-ray dose (0.2 MGy) | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Carpentier, P, Royant, A, Weik, M, Bourgeois, D. | | Deposit date: | 2010-04-14 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Raman Assisted Crystallography Reveals a Mechanism of X-Ray Induced Reversible Disulfide Radical Formation
Structure, 18, 2010
|
|
2XP8
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 4-(MORPHOLIN-4-YLCARBONYL)-2-PHENYL-1H-IMIDAZOLE-5-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XCC
 
 | | Crystal structure of PcrH from Pseudomonas aeruginosa | | Descriptor: | REGULATORY PROTEIN PCRH | | Authors: | Job, V, Mattei, P.-J, Lemaire, D, Attree, I, Dessen, A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of Chaperone Recognition of Type III Secretion System Minor Translocator Proteins.
J.Biol.Chem., 285, 2010
|
|
2XPN
 
 | | Crystal structure of a Spt6-Iws1(Spn1) complex from Encephalitozoon cuniculi, Form I | | Descriptor: | BROMIDE ION, CHROMATIN STRUCTURE MODULATOR, IWS1 | | Authors: | Diebold, M.-L, Koch, M, Cura, V, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-27 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
2XR0
 
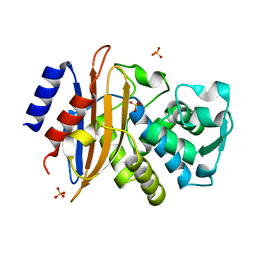 | | Room temperature X-ray structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | Descriptor: | SULFATE ION, TOHO-1 BETA-LACTAMASE | | Authors: | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
2XE7
 
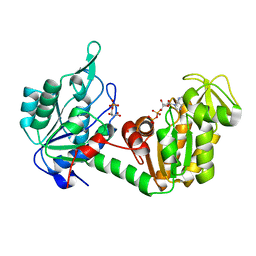 | | The complete reaction cycle of human phosphoglycerate kinase: The open ternary complex with 3PG and ADP | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, PHOSPHOGLYCERATE KINASE 1 | | Authors: | Cliff, M.J, Baxter, N.J, Blackburn, G.M, Merli, A, Vas, M, Waltho, J.P, Bowler, M.W. | | Deposit date: | 2010-05-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Spring Loaded Release Mechanism Regulates Domain Movement and Catalysis in Phosphoglycerate Kinase.
J.Biol.Chem., 286, 2011
|
|
2XRD
 
 | | Structure of the N-terminal four domains of the complement regulator Rat Crry | | Descriptor: | COMPLEMENT REGULATORY PROTEIN CRRY | | Authors: | Leath, K.J, Roversi, P, Johnson, S, Morgan, B.P, Lea, S.M. | | Deposit date: | 2010-09-14 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of the Rat Complement Regulator Crry.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
8RDJ
 
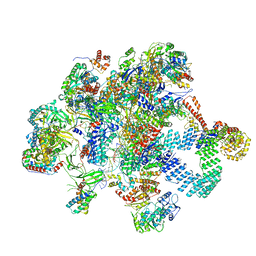 | | Plastid-encoded RNA polymerase transcription elongation complex (Integrated model) | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
