2PPB
 
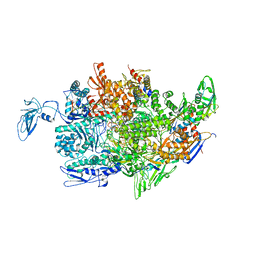 | | Crystal structure of the T. thermophilus RNAP polymerase elongation complex with the ntp substrate analog and antibiotic streptolydigin | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA (5'-D(*AP*AP*CP*GP*CP*CP*AP*GP*AP*CP*AP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*TP*GP*TP*CP*TP*GP*GP*CP*GP*TP*TP*CP*GP*CP*GP*CP*GP*CP*CP*G)-3'), ... | | Authors: | Vassylyev, D.G, Vassylyeva, M.N, Artsimovitch, I, Landick, R. | | Deposit date: | 2007-04-28 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for substrate loading in bacterial RNA polymerase.
Nature, 448, 2007
|
|
5OHQ
 
 | |
8JLB
 
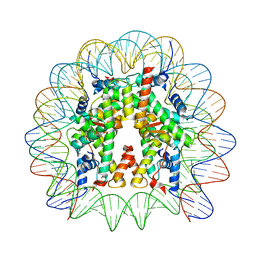 | | Cryo-EM structure of the 145 bp human nucleosome containing H3.2 C110A mutant | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
8JLD
 
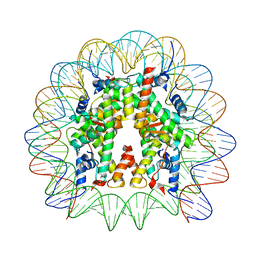 | | Cryo-EM structure of the 145 bp human nucleosome containing acetylated H3 tail | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
2O5J
 
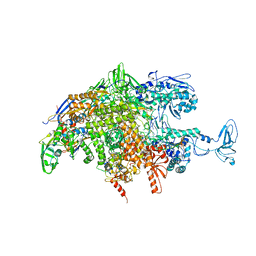 | |
6VJS
 
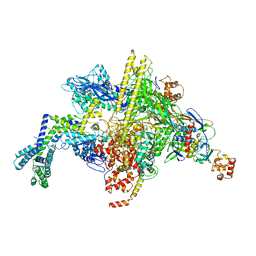 | | Escherichia coli RNA polymerase and ureidothiophene-2-carboxylic acid complex | | Descriptor: | 3-{[benzyl(ethyl)carbamoyl]amino}-5-(4-phenoxyphenyl)thiophene-2-carboxylic acid, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Murakami, K.S, Molodtsov, V. | | Deposit date: | 2020-01-17 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.02 Å) | | Cite: | Evaluation of Bacterial RNA Polymerase Inhibitors in a Staphylococcus aureus -Based Wound Infection Model in SKH1 Mice.
Acs Infect Dis., 6, 2020
|
|
3QQC
 
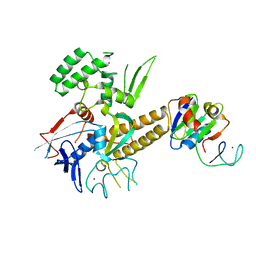 | | Crystal structure of archaeal Spt4/5 bound to the RNAP clamp domain | | Descriptor: | DNA-directed RNA polymerase subunit b, DNA-directed RNA polymerase subunit a', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Martinez-Rucobo, F.W. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity.
Embo J., 30, 2011
|
|
2EC0
 
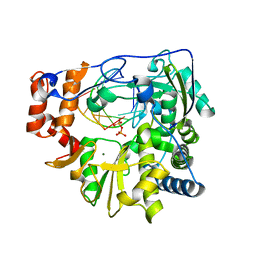 | | RNA-dependent RNA polymerase of foot-and-mouth disease virus in complex with a template-primer RNA and ATP | | Descriptor: | 5'-R(*GP*GP*GP*CP*CP*CP*A)-3', 5'-R(P*AP*UP*GP*GP*GP*CP*CP*C)-3', MAGNESIUM ION, ... | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Sequential structures provide insights into the fidelity of RNA replication
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8JL9
 
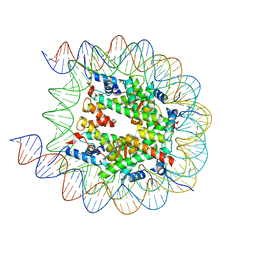 | | Cryo-EM structure of the human nucleosome with scFv | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
8TO6
 
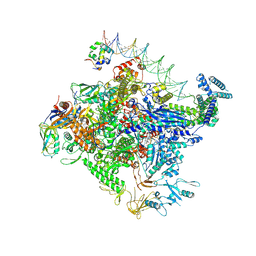 | | Escherichia coli RNA polymerase unwinding intermediate (I1d) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8TOM
 
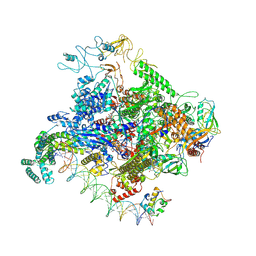 | | Escherichia coli RNA polymerase closed complex intermediate at the lambda PR promoter | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8TOE
 
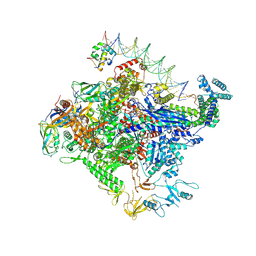 | | Escherichia coli RNA polymerase unwinding intermediate (I1c) at the lambda PR promoter | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-03 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8TO1
 
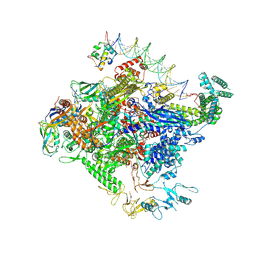 | | Escherichia coli RNA polymerase unwinding intermediate (I1a) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-03 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8TO8
 
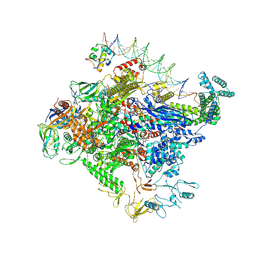 | | Escherichia coli RNA polymerase unwinding intermediate (I1b) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-03 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4YGX
 
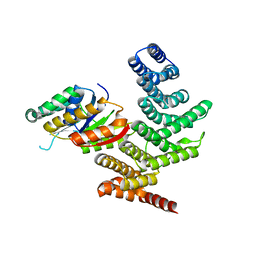 | |
3AYH
 
 | |
1H38
 
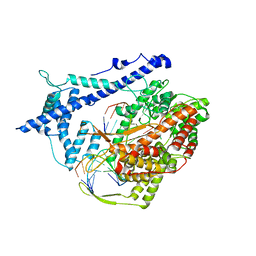 | | Structure of a T7 RNA polymerase elongation complex at 2.9A resolution | | Descriptor: | 5'-D(*GP*GP*GP*AP*AP*TP*CP*GP*AP*CP *AP*TP*CP*GP*CP*CP*GP*C)-3', 5'-D(*GP*TP*CP*GP*AP*TP*TP*CP*CP*CP)-3', 5'-R(*AP*AP*CP*UP*GP*CP*GP*GP*CP*GP *AP*U)-3', ... | | Authors: | Tahirov, T.H, Temyakov, D, Anikin, M, Patlan, V, McAllister, W.T, Vassylyev, D.G, Yokoyama, S. | | Deposit date: | 2002-08-24 | | Release date: | 2002-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a T7 RNA Polymerase Elongation Complex at 2.9 A Resolution
Nature, 420, 2002
|
|
8JLA
 
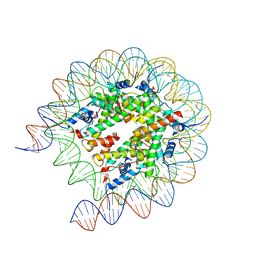 | | Cryo-EM structure of the human nucleosome lacking N-terminal region of H2A, H2B, H3, and H4 | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
7MKP
 
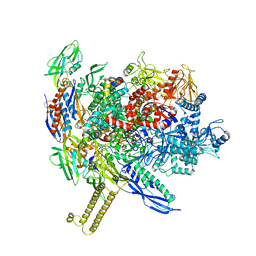 | | Escherichia coli RNA polymerase core enzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural basis of RNA polymerase recycling by the Swi2/Snf2 family of ATPase RapA in Escherichia coli.
J.Biol.Chem., 297, 2021
|
|
7MKO
 
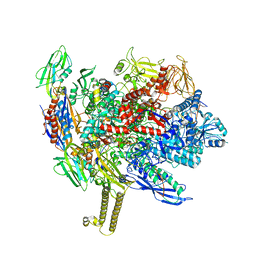 | | Escherichia coli RNA polymerase elongation complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of RNA polymerase recycling by the Swi2/Snf2 family of ATPase RapA in Escherichia coli.
J.Biol.Chem., 297, 2021
|
|
1HQM
 
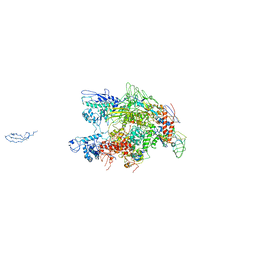 | | CRYSTAL STRUCTURE OF THERMUS AQUATICUS CORE RNA POLYMERASE-INCLUDES COMPLETE STRUCTURE WITH SIDE-CHAINS (EXCEPT FOR DISORDERED REGIONS)-FURTHER REFINED FROM ORIGINAL DEPOSITION-CONTAINS ADDITIONAL SEQUENCE INFORMATION | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Minakhin, L, Bhagat, S, Brunning, A, Campbell, E.A, Darst, S.A, Ebright, R.H, Severinov, K. | | Deposit date: | 2000-12-18 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1NB7
 
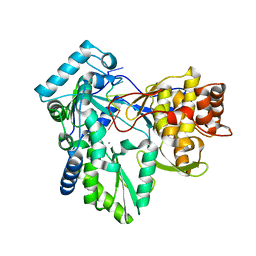 | | HC-J4 RNA polymerase complexed with short RNA template strand | | Descriptor: | 5'-R(*UP*UP*UP*U)-3', MANGANESE (II) ION, polyprotein | | Authors: | O'Farrell, D.J, Trowbridge, R, Rowlands, D.J, Jaeger, J. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation.
J.Mol.Biol., 326, 2003
|
|
1YV2
 
 | | Hepatitis C virus NS5B RNA-dependent RNA Polymerase genotype 2a | | Descriptor: | GLYCEROL, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the RNA-dependent RNA Polymerase Genotype 2a of Hepatitis C Virus Reveal Two Conformations and Suggest Mechanisms of Inhibition by Non-nucleoside Inhibitors
J.Biol.Chem., 280, 2005
|
|
2A69
 
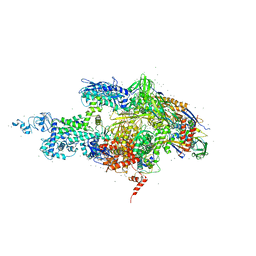 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic rifapentin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
2A68
 
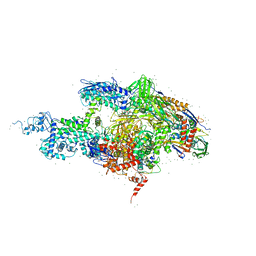 | | Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic rifabutin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
