1RT7
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC84 | | Descriptor: | 1-METHYL ETHYL 1-CHLORO-5-[[(5,6DIHYDRO-2-METHYL-1,4-OXATHIIN-3-YL)CARBONYL]AMINO]BENZOATE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
2Q8M
 
 | | T-like Fructose-1,6-bisphosphatase from Escherichia coli with AMP, Glucose 6-phosphate, and Fructose 1,6-bisphosphate bound | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Hines, J.K, Kruesel, C.E, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-06-11 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Inhibited Fructose-1,6-bisphosphatase from Escherichia coli: DISTINCT ALLOSTERIC INHIBITION SITES FOR AMP AND GLUCOSE 6-PHOSPHATE AND THE CHARACTERIZATION OF A GLUCONEOGENIC SWITCH.
J.Biol.Chem., 282, 2007
|
|
4JPG
 
 | | 2-((1H-benzo[d]imidazol-1-yl)methyl)-4H-pyrido[1,2-a]pyrimidin-4-ones as Novel PKM2 Activators | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-(1H-benzimidazol-1-ylmethyl)-4H-pyrido[1,2-a]pyrimidin-4-one, Pyruvate kinase isozymes M1/M2 | | Authors: | Greasley, S.E, Hickey, M, Phonephaly, H, Cronin, C. | | Deposit date: | 2013-03-19 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery of 2-((1H-benzo[d]imidazol-1-yl)methyl)-4H-pyrido[1,2-a]pyrimidin-4-ones as novel PKM2 activators.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4B3O
 
 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, 5'-D(*CP*GP*TP*AP*TP*GP*CP*CP*TP*AP*TP*AP*GP*TP *TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3', 5'-R(*AP*UP*GP*AP*3DRP*GP*GP*CP*CP*AP*CP*AP*AP*UP*AP *AP*CP*UP*AP*UP*AP*GP*GP*CP*AP*UP*A)-3', ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
3QEF
 
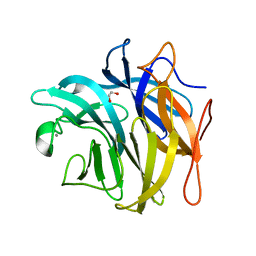 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
4JBY
 
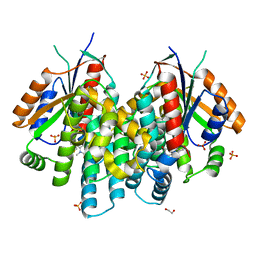 | | Crystal structure of thymidine kinase from Herpes simplex virus type 1 in complex with F-SK78 | | Descriptor: | 1,2-ETHANEDIOL, 6-[(1Z)-3-fluoro-2-(hydroxymethyl)prop-1-en-1-yl]-1,5-dimethylpyrimidine-2,4(1H,3H)-dione, SULFATE ION, ... | | Authors: | Pernot, L, Novakovic, I, Westermaier, Y, Perozzo, R, Raic-Malic, S, Scapozza, L. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New thymine derivatives as HSV1-TK ligand for the development of PET imaging tracer
To be Published
|
|
1PMH
 
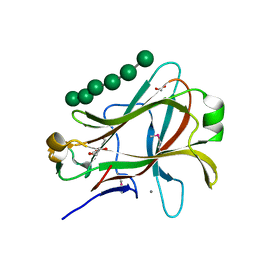 | | Crystal structure of Caldicellulosiruptor saccharolyticus CBM27-1 in complex with mannohexaose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, beta-1,4-mannanase, ... | | Authors: | Roske, Y, Sunna, A, Heinemann, U. | | Deposit date: | 2003-06-11 | | Release date: | 2004-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | High-resolution crystal structures of Caldicellulosiruptor strain Rt8B.4 carbohydrate-binding module CBM27-1 and its complex with mannohexaose.
J.Mol.Biol., 340, 2004
|
|
2PE1
 
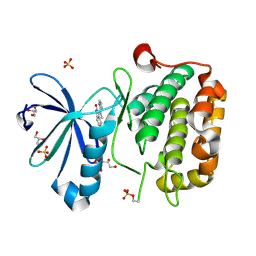 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1 (PDK1) {2-Oxo-3-[1-(1H-pyrrol-2-yl)-eth-(Z)-ylidene]-2,3-dihydro-1H-indol-5-yl}-urea {BX-517} COMPLEX | | Descriptor: | 1-{2-OXO-3-[(1R)-1-(1H-PYRROL-2-YL)ETHYL]-2H-INDOL-5-YL}UREA, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, ... | | Authors: | Whitlow, M, Adler, M. | | Deposit date: | 2007-04-01 | | Release date: | 2007-06-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Indolinone based phosphoinositide-dependent kinase-1 (PDK1) inhibitors. Part 1: Design, synthesis and biological activity.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1AK4
 
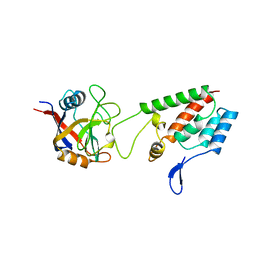 | | HUMAN CYCLOPHILIN A BOUND TO THE AMINO-TERMINAL DOMAIN OF HIV-1 CAPSID | | Descriptor: | CYCLOPHILIN A, HIV-1 CAPSID | | Authors: | Hill, C.P, Gamble, T.R, Vajdos, F.F, Worthylake, D.K, Sundquist, W.I. | | Deposit date: | 1997-05-28 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid.
Cell(Cambridge,Mass.), 87, 1996
|
|
1B6L
 
 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 4 | | Descriptor: | 1-[2-(8-CARBAMOYLMETHYL-6,9-DIOXO-2-OXA-7,10-DIAZA-BICYCLO[11.2.2]HEPTADECA- 1(16),13(17),14-TRIEN-11-YL)-2-HYDROXY-ETHYL]-PIPERIDINE-2-CARBOXYLIC ACID TERT-BUTYLAMIDE, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
1C72
 
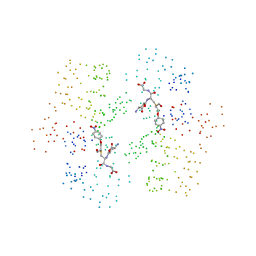 | | TYR115, GLN165 AND TRP209 CONTRIBUTE TO THE 1,2-EPOXY-3-(P-NITROPHENOXY)PROPANE CONJUGATING ACTIVITIES OF GLUTATHIONE S-TRANSFERASE CGSTM1-1 | | Descriptor: | 1-HYDROXY-2-S-GLUTATHIONYL-3-PARA-NITROPHENOXY-PROPANE, PROTEIN (GLUTATHIONE S-TRANSFERASE) | | Authors: | Chern, M.K, Wu, T.C, Hsieh, C.H, Chou, C.C, Liu, L.F, Kuan, I.C, Yeh, Y.H, Hsiao, C.D, Tam, M.F. | | Deposit date: | 2000-02-02 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tyr115, gln165 and trp209 contribute to the 1, 2-epoxy-3-(p-nitrophenoxy)propane-conjugating activity of glutathione S-transferase cGSTM1-1.
J.Mol.Biol., 300, 2000
|
|
1DLT
 
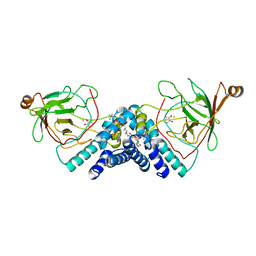 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 WITH BOUND CATECHOL | | Descriptor: | CATECHOL, CATECHOL 1,2-DIOXYGENASE, FE (III) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-12 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1BO5
 
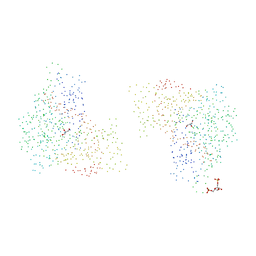 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN ESCHERICHIA COLI GLYCEROL KINASE AND THE ALLOSTERIC REGULATOR FRUCTOSE 1,6-BISPHOSPHATE. | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, PROTEIN (GLYCEROL KINASE) | | Authors: | Ormo, M, Bystrom, C.E, Remington, S.J. | | Deposit date: | 1998-08-10 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a complex of Escherichia coli glycerol kinase and an allosteric effector fructose 1,6-bisphosphate.
Biochemistry, 37, 1998
|
|
2FZZ
 
 | | Factor Xa in complex with the inhibitor 1-(3-amino-1,2-benzisoxazol-5-yl)-6-(2'-(((3r)-3-hydroxy-1-pyrrolidinyl)methyl)-4-biphenylyl)-3-(trifluoromethyl)-1,4,5,6-tetrahydro-7h-pyrazolo[3,4-c]pyridin-7-one | | Descriptor: | 1-(3-AMINO-1,2-BENZISOXAZOL-5-YL)-6-(2'-{[(3R)-3-HYDROXYPYRROLIDIN-1-YL]METHYL}BIPHENYL-4-YL)-3-(TRIFLUOROMETHYL)-1,4,5,6-TETRAHYDRO-7H-PYRAZOLO[3,4-C]PYRIDIN-7-ONE, Coagulation factor X | | Authors: | Alexander, R.S. | | Deposit date: | 2006-02-10 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 1-[3-Aminobenzisoxazol-5'-yl]-3-trifluoromethyl-6-[2'-(3-(R)-hydroxy-N-pyrrolidinyl)methyl-[1,1']-biphen-4-yl]-1,4,5,6-tetrahydropyrazolo-[3,4-c]-pyridin-7-one (BMS-740808) a highly potent, selective, efficacious, and orally bioavailable inhibitor of blood coagulation factor Xa.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2JPR
 
 | | Joint refinement of the HIV-1 CA-NTD in complex with the assembly inhibitor CAP-1 | | Descriptor: | 1-(3-chloro-4-methylphenyl)-3-{2-[({5-[(dimethylamino)methyl]-2-furyl}methyl)thio]ethyl}urea, Gag-Pol polyprotein | | Authors: | Kelly, B.N, Kyere, S, Kinde, I, Tang, C, Howard, B.R, Robinson, H, Sundquist, W.I, Summers, M.F, Hill, C.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein
J.Mol.Biol., 373, 2007
|
|
2LYA
 
 | |
1E48
 
 | |
1E47
 
 | |
2J8F
 
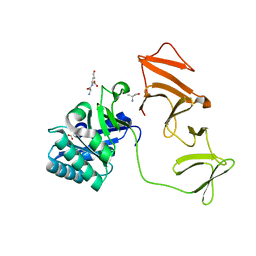 | |
1DOG
 
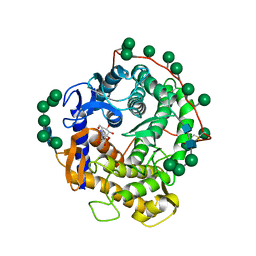 | | REFINED STRUCTURE FOR THE COMPLEX OF 1-DEOXYNOJIRIMYCIN WITH GLUCOAMYLASE FROM (ASPERGILLUS AWAMORI) VAR. X100 TO 2.4 ANGSTROMS RESOLUTION | | Descriptor: | 1-DEOXYNOJIRIMYCIN, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Harris, E, Aleshin, A, Firsov, L, Honzatko, R.B. | | Deposit date: | 1993-01-12 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined structure for the complex of 1-deoxynojirimycin with glucoamylase from Aspergillus awamori var. X100 to 2.4-A resolution.
Biochemistry, 32, 1993
|
|
7A62
 
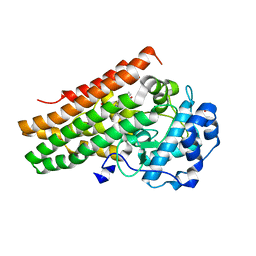 | | Structure of human indoleamine-2,3-dioxygenase 1 (hIDO1) with a complete JK loop | | Descriptor: | CHLORIDE ION, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Mirgaux, M, Wouters, J. | | Deposit date: | 2020-08-24 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43796444 Å) | | Cite: | Influence of the presence of the heme cofactor on the JK-loop structure in indoleamine 2,3-dioxygenase 1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
9EVX
 
 | | cryoEM structure of Photosystem II averaged across S2-S3 states at 1.71 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Graca, A, Zouni, A, Messinger, J, Schroder, W.P. | | Deposit date: | 2024-04-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (1.71 Å) | | Cite: | Cryo-electron microscopy reveals hydrogen positions and water networks in photosystem II.
Science, 384, 2024
|
|
6V52
 
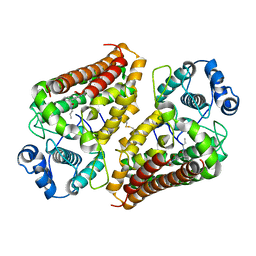 | | IDO1 IN COMPLEX WITH COMPOUND 1 | | Descriptor: | 3-chloro-N-{4-[1-(propylcarbamoyl)cyclobutyl]phenyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Koenig, K.V, Augustin, M.A. | | Deposit date: | 2019-12-03 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Strategic Incorporation of Polarity in Heme-Displacing Inhibitors of Indoleamine-2,3-dioxygenase-1 (IDO1).
Acs Med.Chem.Lett., 11, 2020
|
|
7Z6C
 
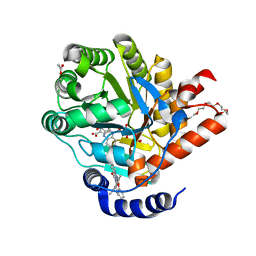 | | Crystal structure of human Dihydroorotate Dehydrogenase in complex with the inhibitor 2-Hydroxy-N-(2-ispropyl-5-methyl-4-phenoxyphenyl)pyrazolo[1,5-a]pyridine-3-carboxamide. | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Alberti, M, Lolli, M.L, Boschi, D, Sainas, S, Rizzi, M, Ferraris, D.M, Miggiano, R. | | Deposit date: | 2022-03-11 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting Acute Myelogenous Leukemia Using Potent Human Dihydroorotate Dehydrogenase Inhibitors Based on the 2-Hydroxypyrazolo[1,5- a ]pyridine Scaffold: SAR of the Aryloxyaryl Moiety.
J.Med.Chem., 65, 2022
|
|
4UB8
 
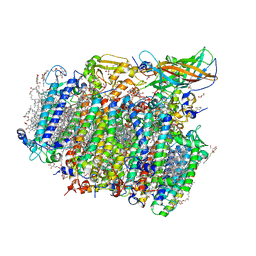 | | Native structure of photosystem II (dataset-2) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
