3QYW
 
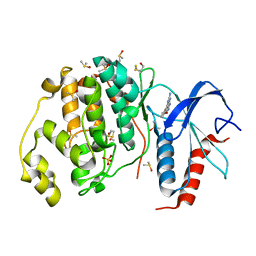 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 6-(3-bromophenyl)-7H-purin-2-amine, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Gelin, M, Pochet, S, Hoh, F, Pirochi, M, Guichou, J.-F, Ferrer, J.-L, Labesse, G. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3HSO
 
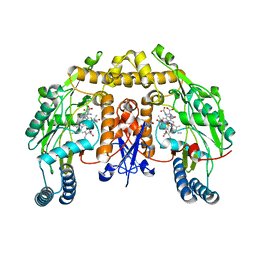 | | Ternary structure of neuronal nitric oxide synthase with NHA and NO bound(1) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-OMEGA-HYDROXY-L-ARGININE, ... | | Authors: | Doukov, T, Li, H, Soltis, M, Poulos, T.L. | | Deposit date: | 2009-06-10 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Single crystal structural and absorption spectral characterizations of nitric oxide synthase complexed with N(omega)-hydroxy-L-arginine and diatomic ligands.
Biochemistry, 48, 2009
|
|
3R1H
 
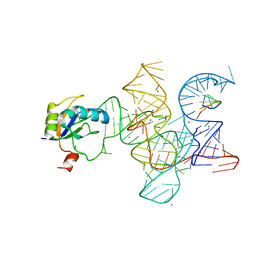 | | Crystal structure of the Class I ligase ribozyme-substrate preligation complex, C47U mutant, Ca2+ bound | | Descriptor: | 5'-R(*UP*CP*CP*AP*GP*UP*A)-3', CALCIUM ION, Class I ligase ribozyme, ... | | Authors: | Shechner, D.M, Bartel, D.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structural basis of RNA-catalyzed RNA polymerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4HHV
 
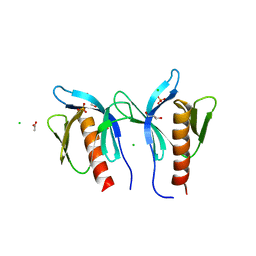 | | Crystal structure of ceramide transfer protein pleckstrin homology domain | | Descriptor: | ACETATE ION, CHLORIDE ION, Collagen type IV alpha-3-binding protein, ... | | Authors: | Prashek, J, Truong, T, Yao, X. | | Deposit date: | 2012-10-10 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Pleckstrin Homology Domain from the Ceramide Transfer Protein: Implications for Conformational Change upon Ligand Binding.
Plos One, 8, 2013
|
|
3WJO
 
 | | Crystal structure of Octaprenyl Pyrophosphate synthase from Escherichia coli with isopentenyl pyrophosphate (IPP) | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Octaprenyl diphosphate synthase | | Authors: | Han, X, Chen, C.C, Kuo, C.J, Huang, C.H, Zheng, Y, Ko, T.P, Zhu, Z, Feng, X, Oldfield, E, Liang, P.H, Guo, R.T, Ma, Y.H. | | Deposit date: | 2013-10-12 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of ligand-bound octaprenyl pyrophosphate synthase from Escherichia coli reveal the catalytic and chain-length determining mechanisms.
Proteins, 83, 2015
|
|
3FO6
 
 | | Crystal structure of guanine riboswitch bound to 6-O-methylguanine | | Descriptor: | 6-O-methylguanine, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Gilbert, S.D, Reyes, F.E, Batey, R.T. | | Deposit date: | 2008-12-28 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adaptive ligand binding by the purine riboswitch in the recognition of Guanine and adenine analogs.
Structure, 17, 2009
|
|
5UTD
 
 | | Sperm whale myoglobin V68A/I107Y with nitrite | | Descriptor: | Myoglobin, NITRITE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2017-02-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
2H26
 
 | | human CD1b in complex with endogenous phosphatidylcholine and spacer | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Garcia-Alles, L.F, Maveyraud, L, Vallina, A.T, Guillet, V, Mourey, L. | | Deposit date: | 2006-05-18 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b.
Embo J., 25, 2006
|
|
3WJK
 
 | | Crystal structure of Octaprenyl Pyrophosphate synthase from Escherichia coli | | Descriptor: | Octaprenyl diphosphate synthase | | Authors: | Han, X, Chen, C.C, Kuo, C.J, Huang, C.H, Zheng, Y, Ko, T.P, Zhu, Z, Feng, X, Oldfield, E, Liang, P.H, Guo, R.T, Ma, Y.H. | | Deposit date: | 2013-10-11 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of ligand-bound octaprenyl pyrophosphate synthase from Escherichia coli reveal the catalytic and chain-length determining mechanisms.
Proteins, 83, 2015
|
|
1JOJ
 
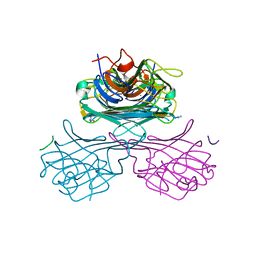 | | CONCANAVALIN A-HEXAPEPTIDE COMPLEX | | Descriptor: | CALCIUM ION, Concanavalin-Br, HEXAPEPTIDE, ... | | Authors: | Jain, D, Kaur, K, Salunke, D.M. | | Deposit date: | 2001-07-30 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Enhanced binding of a rationally designed peptide ligand of concanavalin a arises from improved geometrical complementarity.
Biochemistry, 40, 2001
|
|
1JZK
 
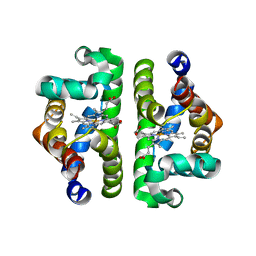 | | Crystal Structure of Scapharca inaequivalvis HbI, I114F mutant (deoxy) | | Descriptor: | GLOBIN I - ARK SHELL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Gibson, Q.H, Cushing, L, Royer Jr, W.E. | | Deposit date: | 2001-09-16 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Restricting the Ligand-Linked Heme Movement in Scapharca Dimeric Hemoglobin Reveals Tight Coupling between Distal and Proximal
Contributions to Cooperativity.
Biochemistry, 40, 2001
|
|
4QR6
 
 | |
2C9W
 
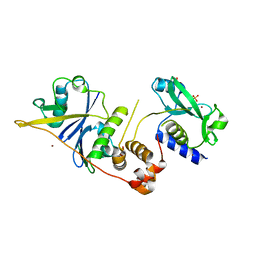 | | CRYSTAL STRUCTURE OF SOCS-2 IN COMPLEX WITH ELONGIN-B AND ELONGIN-C AT 1.9A RESOLUTION | | Descriptor: | NICKEL (II) ION, SULFATE ION, SUPPRESSOR OF CYTOKINE SIGNALING 2, ... | | Authors: | Debreczeni, J.E, Bullock, A, Amos, A, Savitsky, P, Barr, A, Burgess, N, Sundstrom, M, Weigelt, J, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 103, 2006
|
|
3WJN
 
 | | Crystal structure of Octaprenyl Pyrophosphate synthase from Escherichia coli with farnesyl S-thiol-pyrophosphate (FSPP) | | Descriptor: | Octaprenyl diphosphate synthase, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE | | Authors: | Han, X, Chen, C.C, Kuo, C.J, Huang, C.H, Zheng, Y, Ko, T.P, Zhu, Z, Feng, X, Oldfield, E, Liang, P.H, Guo, R.T, Ma, Y.H. | | Deposit date: | 2013-10-12 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of ligand-bound octaprenyl pyrophosphate synthase from Escherichia coli reveal the catalytic and chain-length determining mechanisms.
Proteins, 83, 2015
|
|
8Q2R
 
 | | Crystal structure of YTHDC1 in complex with Compound 3 (ZA_431) | | Descriptor: | 5-chloranyl-~{N},3-dimethyl-1~{H}-pyrazolo[4,3-d]pyrimidin-7-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Zalesak, F, Li, Y, Caflisch, A. | | Deposit date: | 2023-08-03 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Design of a Potent and Selective YTHDC1 Ligand.
J.Med.Chem., 67, 2024
|
|
8Q2Y
 
 | |
8Q3G
 
 | |
8Q2X
 
 | |
8Q4W
 
 | | Crystal structure of YTHDC1 in complex with Compound 40 (CS_3a) | | Descriptor: | 5-chloranyl-3-[(3-chlorophenyl)methyl]-~{N}-methyl-2~{H}-pyrazolo[4,3-d]pyrimidin-7-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Zalesak, F, Caflisch, A. | | Deposit date: | 2023-08-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure-Based Design of a Potent and Selective YTHDC1 Ligand.
J.Med.Chem., 67, 2024
|
|
8Q3A
 
 | |
8Q4N
 
 | |
8VFW
 
 | | Crystal Structure of V113N D-Dopachrome Tautomerase (D-DT) | | Descriptor: | CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VFO
 
 | | Crystal Structure of L117G Variant of D-Dopachrome Tautomerase (D-DT) | | Descriptor: | CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8Q2S
 
 | |
8VG8
 
 | | Crystal Structure of T115A Variant of D-Dopachrome Tautomerase (D-DT) Bound to 4CPPC | | Descriptor: | 4-(3-carboxyphenyl)pyridine-2,5-dicarboxylic acid, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
