7KZU
 
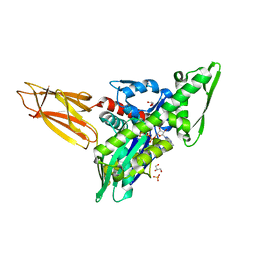 | | Quasi-intermediate state (Q) of a truncated Hsp70 DnaK fused with a substrate peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK fused with substrate peptide,Chaperone protein DnaK fused with substrate peptide, GLYCEROL, ... | | Authors: | Wang, W, Hendrickson, W.A. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Intermediates in allosteric equilibria of DnaK-ATP interactions with substrate peptides
Acta Crystallogr.,Sect.D, 77, 2021
|
|
4H6O
 
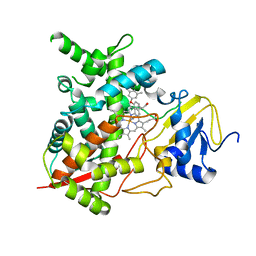 | | Sterol 14-alpha demethylase (CYP51)from Trypanosoma cruzi in complex with the inhibitor NEU321 (1-(3-(4-chloro-3,5-dimethylphenoxy)benzyl)-1H-imidazole | | Descriptor: | 1-[3-(4-chloro-3,5-dimethylphenoxy)benzyl]-1H-imidazole, PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha demethylase | | Authors: | Wawrzak, Z, Hargrove, T.Y, Pollastri, M.P, Lepesheva, G.I. | | Deposit date: | 2012-09-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antitrypanosomal Lead Discovery: Identification of a Ligand-Efficient Inhibitor of Trypanosoma cruzi CYP51 and Parasite Growth.
J.Med.Chem., 56, 2013
|
|
4JTY
 
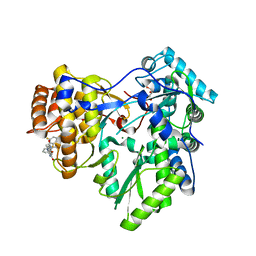 | | Crystal structure of HCV NS5B polymerase with COMPOUND 2 | | Descriptor: | 3-{[4-oxo-1-(2,4,6-trifluorobenzyl)-1,4-dihydroquinazolin-6-yl]oxy}-2-(trifluoromethyl)benzamide, GLYCEROL, Genome polyprotein, ... | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-24 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Dynamics Simulations and Structure-Based Rational Design Lead to Allosteric HCV NS5B Polymerase Thumb Pocket 2 Inhibitor with Picomolar Cellular Replicon Potency.
J.Med.Chem., 57, 2014
|
|
3GMD
 
 | | Structure-based design of 7-Azaindole-pyrrolidines as inhibitors of 11beta-Hydroxysteroid-Dehydrogenase type I | | Descriptor: | 2-methyl-3-{(3S)-1-[(1-pyridin-2-ylcyclopropyl)carbonyl]pyrrolidin-3-yl}-1H-pyrrolo[2,3-b]pyridine, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Valeur, E, Lepifre, F, Roche, D, Christmann-Franck, S, Hillertz, P, Musil, D. | | Deposit date: | 2009-03-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-based design of 7-azaindole-pyrrolidine amides as inhibitors of 11 beta-hydroxysteroid dehydrogenase type I.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4JU2
 
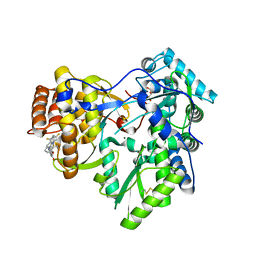 | | Crystal structure of hcv ns5b polymerase in complex with compound 12 | | Descriptor: | 6-[3-(pyridin-2-yl)-2-(trifluoromethyl)phenoxy]-1-(2,4,6-trifluorobenzyl)quinazolin-4(1H)-one, GLYCEROL, Genome polyprotein, ... | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-24 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Dynamics Simulations and Structure-Based Rational Design Lead to Allosteric HCV NS5B Polymerase Thumb Pocket 2 Inhibitor with Picomolar Cellular Replicon Potency.
J.Med.Chem., 57, 2014
|
|
4JTW
 
 | | Crystal structure of HCV NS5B polymerase in complex with coupound 1 | | Descriptor: | 1-(2,4,6-trifluorobenzyl)-6-[2-(trifluoromethyl)phenoxy]quinazolin-4(1H)-one, GLYCEROL, Genome polyprotein, ... | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-24 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Dynamics Simulations and Structure-Based Rational Design Lead to Allosteric HCV NS5B Polymerase Thumb Pocket 2 Inhibitor with Picomolar Cellular Replicon Potency.
J.Med.Chem., 57, 2014
|
|
4JU1
 
 | | Crystal structure of hcv ns5b polymerase in complex with compound 6 | | Descriptor: | 6-[3-([1,3]oxazolo[4,5-b]pyridin-2-yl)-2-(trifluoromethyl)phenoxy]-1-(2,4,6-trifluorobenzyl)quinazolin-4(1H)-one, GLYCEROL, Genome polyprotein, ... | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-24 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Dynamics Simulations and Structure-Based Rational Design Lead to Allosteric HCV NS5B Polymerase Thumb Pocket 2 Inhibitor with Picomolar Cellular Replicon Potency.
J.Med.Chem., 57, 2014
|
|
4JTZ
 
 | | Crystal structure of hcv ns5b polymerase in complex with compound 4 | | Descriptor: | 3-{[4-oxo-1-(2,4,6-trifluorobenzyl)-1,4-dihydroquinazolin-6-yl]oxy}-N-(pyridin-3-yl)-2-(trifluoromethyl)benzamide, GLYCEROL, Genome polyprotein, ... | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-24 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Dynamics Simulations and Structure-Based Rational Design Lead to Allosteric HCV NS5B Polymerase Thumb Pocket 2 Inhibitor with Picomolar Cellular Replicon Potency.
J.Med.Chem., 57, 2014
|
|
5LYG
 
 | | CRYSTAL STRUCTURE OF INTRACELLULAR B30.2 DOMAIN OF BTN3A1 BOUND TO MALONATE | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, MALONATE ION | | Authors: | Mohammed, F, Baker, A.T, Salim, M, Willcox, B.E. | | Deposit date: | 2016-09-28 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | BTN3A1 Discriminates gamma delta T Cell Phosphoantigens from Nonantigenic Small Molecules via a Conformational Sensor in Its B30.2 Domain.
ACS Chem. Biol., 12, 2017
|
|
5LYK
 
 | | CRYSTAL STRUCTURE OF INTRACELLULAR B30.2 DOMAIN OF BTN3A1 BOUND TO CITRATE | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, CITRATE ANION | | Authors: | Mohammed, F, Baker, A.T, Salim, M, Willcox, B.E. | | Deposit date: | 2016-09-28 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | BTN3A1 Discriminates gamma delta T Cell Phosphoantigens from Nonantigenic Small Molecules via a Conformational Sensor in Its B30.2 Domain.
ACS Chem. Biol., 12, 2017
|
|
8H73
 
 | | Crystal structure of antibody scFv against M2e Influenza peptide | | Descriptor: | CITRATE ANION, Single Chain Variable Fragment | | Authors: | Kumar, U, Madni, Z.K, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A structure and knowledge-based combinatorial approach to engineering universal scFv antibodies against influenza M2 protein.
J.Biomed.Sci., 30, 2023
|
|
8H3B
 
 | | Crystal structure of antibody scFv against M2e Influenza peptide | | Descriptor: | GLYCEROL, Single Chain Variable Fragment | | Authors: | Kumar, U, Madni, Z.K, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-10-08 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A structure and knowledge-based combinatorial approach to engineering universal scFv antibodies against influenza M2 protein.
J.Biomed.Sci., 30, 2023
|
|
8H3C
 
 | | Crystal structure of M2e Influenza peptide in complex with antibody scFv | | Descriptor: | Matrix protein 2, Single Chain Variable Fragment | | Authors: | Kumar, U, Madni, Z.K, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-10-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | A structure and knowledge-based combinatorial approach to engineering universal scFv antibodies against influenza M2 protein.
J.Biomed.Sci., 30, 2023
|
|
3DQS
 
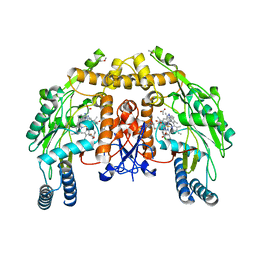 | | Structure of endothelial NOS heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2008-07-09 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
3DRR
 
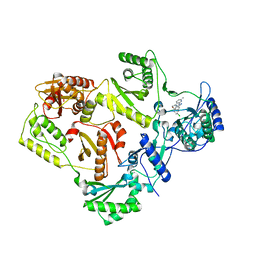 | | HIV reverse transcriptase Y181C mutant in complex with inhibitor R8e | | Descriptor: | 3-{5-[(6-amino-1H-pyrazolo[3,4-b]pyridin-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Yan, Y. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of 3-{5-[(6-Amino-1H-pyrazolo[3,4-b]pyridine-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile (MK-4965): A Potent, Orally Bioavailable HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitor with Improved Potency against Key Mutant Viruses.
J.Med.Chem., 51, 2008
|
|
2WFY
 
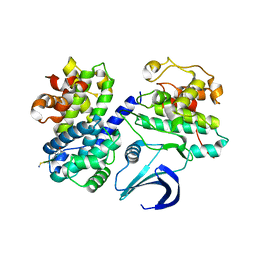 | | Truncation and Optimisation of Peptide Inhibitors of CDK2, Cyclin A Through Structure Guided Design | | Descriptor: | ARG-ARG-B3L-PHE, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2 | | Authors: | Kontopidis, G, Andrews, M.J, McInnes, C, Plater, A, Innes, L, Renachowski, S, Cowan, A, Fischer, P.M. | | Deposit date: | 2009-04-15 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Truncation and Optimisation of Peptide Inhibitors of Cyclin-Dependent Kinase 2-Cyclin a Through Structure-Guided Design.
Chemmedchem, 4, 2009
|
|
2WHB
 
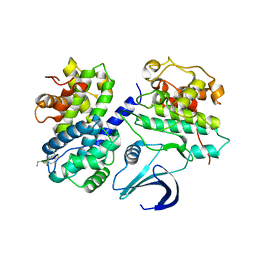 | | Truncation and Optimisation of Peptide Inhibitors of CDK2, Cyclin A Through Structure Guided Design | | Descriptor: | ARG-ARG-L3O-PFF, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2 | | Authors: | Kontopidis, G, Andrews, M.J, McInnes, C, Plater, A, Innes, L, Renachowski, S, Cowan, A, Fischer, P.M. | | Deposit date: | 2009-05-03 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Truncation and optimisation of peptide inhibitors of cyclin-dependent kinase 2-cyclin a through structure-guided design.
Chemmedchem, 4, 2009
|
|
2WEV
 
 | | Truncation and Optimisation of Peptide Inhibitors of CDK2, Cyclin A Through Structure Guided Design | | Descriptor: | ARG-ARG-B3L-MEA, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, ... | | Authors: | Kontopidis, G, Andrews, M.J, McInnes, C, Plater, A, Innes, L, Renachowski, S, Cowan, A, Fischer, P.M. | | Deposit date: | 2009-04-01 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Truncation and Optimisation of Peptide Inhibitors of Cyclin-Dependent Kinase 2-Cyclin a Through Structure-Guided Design.
Chemmedchem, 4, 2009
|
|
3DQR
 
 | | Structure of neuronal NOS D597N/M336V mutant heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-aminopyridin-2"-yl)methyl]pyrrolidin-3'-yl}ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-aminopyridin-2-yl)methyl]pyrrolidin-3-yl}ethane-1,2-diamine, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2008-07-09 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
3DQT
 
 | | Structure of endothelial NOS heme domain in complex with a inhibitor (+-)-N1-{trans-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2008-07-09 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
3DRS
 
 | | HIV reverse transcriptase K103N mutant in complex with inhibitor R8D | | Descriptor: | 3-chloro-5-[2-chloro-5-(1H-pyrazolo[3,4-b]pyridin-3-ylmethoxy)phenoxy]benzonitrile, Reverse transcriptase/ribonuclease H, p66 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Discovery of 3-{5-[(6-Amino-1H-pyrazolo[3,4-b]pyridine-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile (MK-4965): A Potent, Orally Bioavailable HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitor with Improved Potency against Key Mutant Viruses.
J.Med.Chem., 51, 2008
|
|
3DRP
 
 | | HIV reverse transcriptase in complex with inhibitor R8e | | Descriptor: | 3-{5-[(6-amino-1H-pyrazolo[3,4-b]pyridin-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 3-{5-[(6-Amino-1H-pyrazolo[3,4-b]pyridine-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile (MK-4965): A Potent, Orally Bioavailable HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitor with Improved Potency against Key Mutant Viruses.
J.Med.Chem., 51, 2008
|
|
3H23
 
 | |
3H2E
 
 | | Structural Studies of Pterin-Based Inhibitors of Dihydropteroate Synthase | | Descriptor: | 1,3-dimethyl-2,4,7-trioxo-1,2,3,4,7,8-hexahydropteridine-6-carbaldehyde, Dihydropteroate synthase, SULFATE ION | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2009-04-14 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of pterin-based inhibitors of dihydropteroate synthase.
J.Med.Chem., 53, 2010
|
|
3H21
 
 | | Structural Studies of Pterin-Based Inhibitors of Dihydropteroate Synthase | | Descriptor: | (2R)-2-(7-amino-1-methyl-4,5-dioxo-1,4,5,6-tetrahydropyrimido[4,5-c]pyridazin-3-yl)propanoic acid, Dihydropteroate synthase, SULFATE ION | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2009-04-14 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural studies of pterin-based inhibitors of dihydropteroate synthase.
J.Med.Chem., 53, 2010
|
|
