1UXV
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXU
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
7PSM
 
 | | S. cerevisiae Atm1 in MSP1D1 nanodiscs with bound AMP-PNP and Mg2+ | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, Iron-sulfur clusters transporter ATM1, mitochondrial, ... | | Authors: | Ellinghaus, T.L, Kuehlbrandt, W. | | Deposit date: | 2021-09-23 | | Release date: | 2021-12-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Conformational changes in the yeast mitochondrial ABC transporter Atm1 during the transport cycle.
Sci Adv, 7, 2021
|
|
7PSN
 
 | | S. cerevisiae Atm1 in MSP1E3D1 nanodiscs with bound AMP-PNP and Mg2+ | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, Iron-sulfur clusters transporter ATM1, mitochondrial, ... | | Authors: | Ellinghaus, T.L, Kuehlbrandt, W. | | Deposit date: | 2021-09-23 | | Release date: | 2021-12-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational changes in the yeast mitochondrial ABC transporter Atm1 during the transport cycle.
Sci Adv, 7, 2021
|
|
2W4Z
 
 | | Caulobacter bacteriophage 5 | | Descriptor: | ADENOSINE MONOPHOSPHATE, CALCIUM ION, CAULOBACTER BACTERIOPHAGE 5 | | Authors: | Plevka, P, Kazaks, A, Dishlers, A, Liljas, L, Tars, K. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Structure of Bacteriophage Phicb5 Reveals a Role of the RNA Genome and Metal Ions in Particle Stability and Assembly.
J.Mol.Biol., 391, 2009
|
|
2FPL
 
 | | RadA recombinase in complex with AMP-PNP and low concentration of K+ | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Methanococcus Voltae Rada in Complex with Adp: hydrolysis-induced conformational change
Biochemistry, 44, 2005
|
|
2FPM
 
 | | RadA recombinase in complex with AMP-PNP and high concentration of K+ | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Methanoccocus Voltae Rada in Complex with Adp: hydrolysis-induced conformational change
Biochemistry, 44, 2005
|
|
5ZK1
 
 | | Crystal Structure of the CRTC2(SeMet)-CREB-CRE complex | | Descriptor: | CREB-regulated transcription coactivator 2, Cyclic AMP-responsive element-binding protein 1, DNA (5'-D(*CP*TP*TP*GP*GP*CP*TP*GP*AP*CP*GP*TP*CP*AP*GP*CP*CP*AP*AP*G)-3'), ... | | Authors: | Xiang, S, Zhai, L, Valencia-Swain, J. | | Deposit date: | 2018-03-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Insights into the CRTC2-CREB Complex Assembly on CRE.
J. Mol. Biol., 430, 2018
|
|
5ZKO
 
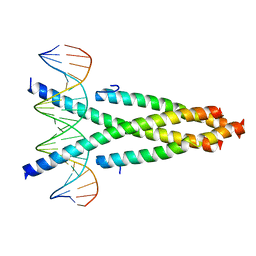 | | Crystal structure of the CRTC2-CREB-CRE complex | | Descriptor: | CREB-regulated transcription coactivator 2, Cyclic AMP-responsive element-binding protein 1, DNA (5'-D(*CP*TP*TP*GP*GP*CP*TP*GP*AP*CP*GP*TP*CP*AP*GP*CP*CP*AP*AP*G)-3') | | Authors: | Xiang, S, Zhai, L, Valecia-Swain, J. | | Deposit date: | 2018-03-24 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Insights into the CRTC2-CREB Complex Assembly on CRE.
J. Mol. Biol., 430, 2018
|
|
2FSH
 
 | |
1UXN
 
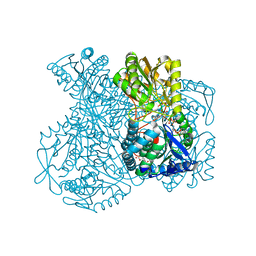 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-02-27 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
2C1B
 
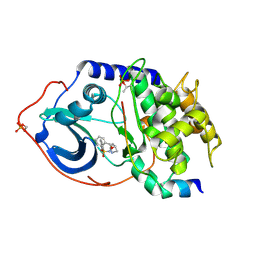 | | Structure of cAMP-dependent protein kinase complexed with (4R,2S)-5'-(4-(4-Chlorobenzyloxy)pyrrolidin-2-ylmethanesulfonyl)isoquinoline | | Descriptor: | (4R,2S)-5'-(4-(4-CHLOROBENZYLOXY)PYRROLIDIN-2-YLMETHANESULFONYL)ISOQUINOLINE, CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR | | Authors: | Collins, I, Caldwell, J, Fonseca, T, Donald, A, Bavetsias, V, Hunter, L.J, Garrett, M.D, Rowlands, M.G, Aherne, G.W, Davies, T.G, Berdini, V, Woodhead, S.J, Seavers, L.C.A, Wyatt, P.G, Workman, P, McDonald, E. | | Deposit date: | 2005-09-12 | | Release date: | 2005-11-02 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of isoquinoline-5-sulfonamide inhibitors of protein kinase B.
Bioorg. Med. Chem., 14, 2006
|
|
5F2R
 
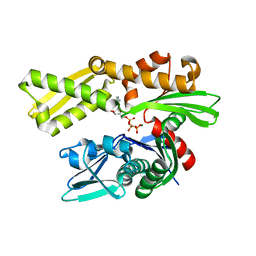 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with AMP-PCP | | Descriptor: | 78 kDa glucose-regulated protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-12-02 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
7WIV
 
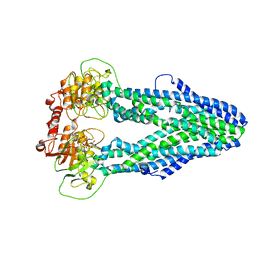 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in complex with an AMP-PNP | | Descriptor: | Mycobactin import ATP-binding/permease protein IrtA, Mycobactin import ATP-binding/permease protein IrtB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
3J4R
 
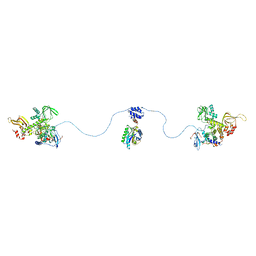 | |
3C0H
 
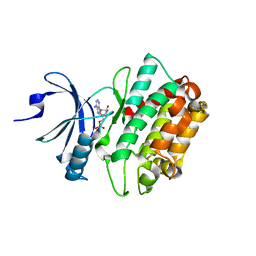 | | CASK CaM-Kinase Domain- AMPPNP complex, P1 form | | Descriptor: | ADENOSINE MONOPHOSPHATE, Peripheral plasma membrane protein CASK | | Authors: | Wahl, M.C. | | Deposit date: | 2008-01-20 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CASK Functions as a Mg2+-independent neurexin kinase
Cell(Cambridge,Mass.), 133, 2008
|
|
7B5U
 
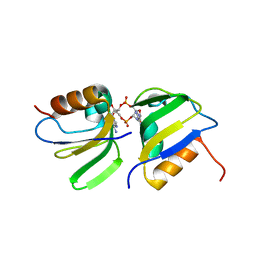 | | RCK_C domain dimer of S.agalactiae BusR in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, GntR family transcriptional regulator | | Authors: | Bandera, A.M, Witte, G. | | Deposit date: | 2020-12-07 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res., 49, 2021
|
|
5TVA
 
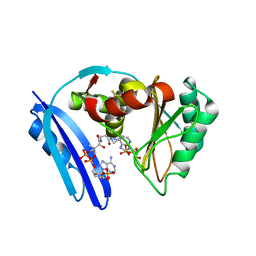 | | A. aeolicus BioW with AMP and CoA | | Descriptor: | 6-carboxyhexanoate--CoA ligase, ADENOSINE MONOPHOSPHATE, COENZYME A | | Authors: | Estrada, P, Manandhar, M, Dong, S.-H, Deveryshetty, J, Agarwal, V, Cronan, J.E, Nair, S.K. | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-07 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes.
Nat. Chem. Biol., 13, 2017
|
|
7R7E
 
 | |
5X0G
 
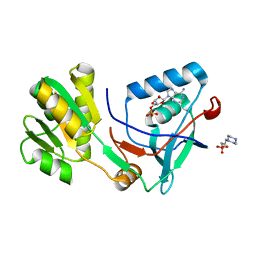 | | Free serine kinase (E30A mutant) in complex with ADP | | Descriptor: | 2-hydroxy-3-[4-(2-hydroxy-3-sulfopropyl)piperazin-1-yl]propane-1-sulfonic acid, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2017-01-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Study on the Reaction Mechanism of a Free Serine Kinase Involved in Cysteine Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
7CCR
 
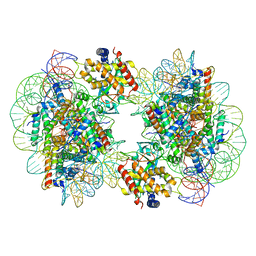 | | Structure of the 2:2 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | Authors: | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
7CCQ
 
 | | Structure of the 1:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | Authors: | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
1CGP
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP)/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*GP*TP*GP*TP*GP*AP*CP*AP*TP*AP*T)-3'), DNA (5'-D(*GP*TP*CP*AP*CP*AP*CP*TP*TP*TP*TP*CP*G)-3'), ... | | Authors: | Schultz, S.C, Shields, G.C, Steitz, T.A. | | Deposit date: | 1991-08-12 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees.
Science, 253, 1991
|
|
4OKE
 
 | |
7LVQ
 
 | | KIF14[391-743] - AMP-PNP closed state class in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF14, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Paydar, M, Dhakal, S, Kwok, B, Sosa, H. | | Deposit date: | 2021-02-26 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of mechano-chemical coupling by the mitotic kinesin KIF14.
Nat Commun, 12, 2021
|
|
