2Z9X
 
 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxyl-L-alanine | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, ALANINE, Aspartate aminotransferase, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
7JX1
 
 | | E. coli TSase complex with a bi-substrate reaction intermediate analog | | Descriptor: | (2S)-2-({4-[({(6R)-2,4-diamino-5-[(1-{(2R,4S,5R)-4-hydroxy-5-[(phosphonooxy)methyl]tetrahydrofuran-2-yl}-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)methyl]-5,6,7,8-tetrahydropyrido[3,2-d]pyrimidin-6-yl}methyl)amino]benzoyl}amino)pentanedioic acid (non-preferred name), N-(4-{[(2,4-diamino-7,8-dihydropyrido[3,2-d]pyrimidin-6-yl)methyl]amino}benzene-1-carbonyl)-L-glutamic acid, PHOSPHATE ION, ... | | Authors: | Finer-Moore, J, Kholodar, S.A, Stroud, R.M, Kohen, A. | | Deposit date: | 2020-08-26 | | Release date: | 2021-04-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Caught in Action: X-ray Structure of Thymidylate Synthase with Noncovalent Intermediate Analog.
Biochemistry, 60, 2021
|
|
1N0Y
 
 | | Crystal Structure of Pb-bound Calmodulin | | Descriptor: | ACETATE ION, CACODYLATE ION, Calmodulin, ... | | Authors: | Wilson, M.A, Brunger, A.T. | | Deposit date: | 2002-10-15 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Domain flexibility in the 1.75 A resolution structure of Pb2+-calmodulin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7KCE
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor compound 2 | | Descriptor: | 5-methyl-2,3-diphenylpyrazolo[1,5-a]pyrimidin-7(4H)-one, CHLORIDE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-05 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
2NSF
 
 | | Crystal structure of the mycothiol-dependent maleylpyruvate isomerase | | Descriptor: | GLYCEROL, Hypothetical protein Cgl3021, SULFATE ION, ... | | Authors: | Chang, W.R, Wang, R. | | Deposit date: | 2006-11-04 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures and Site-directed Mutagenesis of a Mycothiol-dependent Enzyme Reveal a Novel Folding and Molecular Basis for Mycothiol-mediated Maleylpyruvate Isomerization
J.Biol.Chem., 282, 2007
|
|
3W12
 
 | | Insulin receptor ectodomain construct comprising domains L1-CR in complex with high-affinity insulin analogue [D-PRO-B26]-DTI-NH2, alpha-CT peptide(704-719) and FAB 83-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, Insulin B chain, ... | | Authors: | Lawrence, M.C, Smith, B.J, Brzozowsk, A.M. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.301 Å) | | Cite: | How insulin engages its primary binding site on the insulin receptor
Nature, 493, 2013
|
|
2ZEC
 
 | | Potent, Nonpeptide Inhibitors of Human Mast Cell Tryptase | | Descriptor: | 1-[1'-(3-phenylacryloyl)spiro[1-benzofuran-3,4'-piperidin]-5-yl]methanamine, Tryptase beta 2 | | Authors: | Spurlino, J.C, Lewandowski, F, Milligan, C. | | Deposit date: | 2007-12-08 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Potent, nonpeptide inhibitors of human mast cell tryptase. Synthesis and biological evaluation of novel spirocyclic piperidine amide derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5R8A
 
 | | PanDDA analysis group deposition INTERLEUKIN-1 BETA -- Fragment Z1492796719 in complex with INTERLEUKIN-1 BETA | | Descriptor: | Interleukin-1 beta, SULFATE ION, ~{N}-[(3~{R})-1,2,3,4-tetrahydroquinolin-3-yl]ethanamide | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
5R8O
 
 | |
1PX6
 
 | | A folding mutant of human class pi glutathione transferase, created by mutating aspartate 153 of the wild-type protein to asparagine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, Glutathione S-transferase P | | Authors: | Kong, G.K.-W, Polekhina, G, McKinstry, W.J, Parker, M.W, Dragani, B, Aceto, A, Paludi, D, Principe, D.R, Mannervik, B, Stenberg, G. | | Deposit date: | 2003-07-02 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The multi-functional role of a highly conserved aspartic acid residue in glutathione transferase P1-1
To be Published
|
|
3KMG
 
 | |
1MVQ
 
 | | Cratylia mollis lectin (isoform 1) in complex with methyl-alpha-D-mannose | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, lectin, ... | | Authors: | de Souza, G.A, Oliveira, P.S, Trapani, S, Correia, M.T, Oliva, G, Coelho, L.C, Greene, L.J. | | Deposit date: | 2002-09-26 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Amino acid sequence and tertiary structure of Cratylia mollis seed lectin
Glycobiology, 13, 2003
|
|
166L
 
 | | CONTROL OF ENZYME ACTIVITY BY AN ENGINEERED DISULFIDE BOND | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent.
J.Mol.Biol., 246, 1995
|
|
174L
 
 | |
1SVN
 
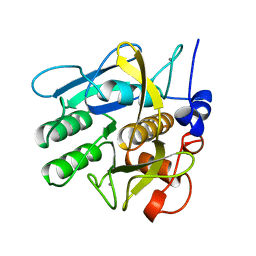 | | SAVINASE | | Descriptor: | CALCIUM ION, SAVINASE (TM) | | Authors: | Betzel, C, Klupsch, S, Papendorf, G, Hastrup, S, Branner, S, Wilson, K.S. | | Deposit date: | 1995-09-01 | | Release date: | 1996-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the alkaline proteinase Savinase from Bacillus lentus at 1.4 A resolution.
J.Mol.Biol., 223, 1992
|
|
1Q5D
 
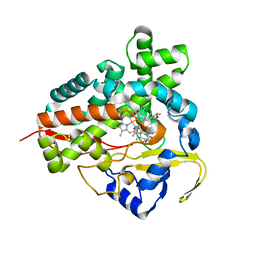 | | Epothilone B-bound Cytochrome P450epoK | | Descriptor: | 7,11-DIHYDROXY-8,8,10,12,16-PENTAMETHYL-3-[1-METHYL-2-(2-METHYL-THIAZOL-4-YL)VINYL]-4,17-DIOXABICYCLO[14.1.0]HEPTADECANE-5,9-DIONE, P450 epoxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
3KDJ
 
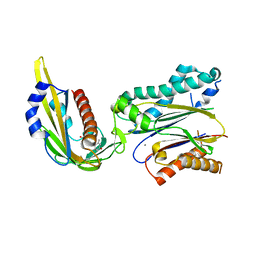 | | Complex structure of (+)-ABA-bound PYL1 and ABI1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, MANGANESE (II) ION, Protein phosphatase 2C 56, ... | | Authors: | Yin, P, Fan, H, Hao, Q, Yuan, X, Yan, N. | | Deposit date: | 2009-10-23 | | Release date: | 2009-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Structural insights into the mechanism of abscisic acid signaling by PYL proteins
Nat.Struct.Mol.Biol., 16, 2009
|
|
1Q5T
 
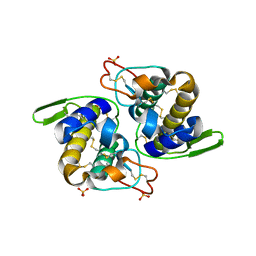 | | Gln48 PLA2 separated from Vipoxin from the venom of Vipera ammodytes meridionalis. | | Descriptor: | Phospholipase A2 inhibitor, SULFATE ION | | Authors: | Georgieva, D.N, Perbandt, M, Rypniewski, W, Hristov, K, Genov, N, Betzel, C. | | Deposit date: | 2003-08-11 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray structure of a snake venom Gln48 phospholipase A2 at 1.9A resolution reveals
anion-binding sites.
Biochem.Biophys.Res.Commun., 316, 2004
|
|
5TA2
 
 | | Discovery of a Potent Cyclophilin Inhibitor (Compound 7) based on Structural Simplification of Sanglifehrin A | | Descriptor: | 11-[(3-hydroxyphenyl)methyl]-18-methoxy-2,17-dimethyl-14-(propan-2-yl)-3-oxa-9,12,15,28-tetraazatricyclo[21.3.1.1~5,9~]octacosa-1(27),21,23,25-tetraene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Steadman, V, Pettit, S, Schmitz, U, Mackman, R.L, Schultz, B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Discovery of Potent Cyclophilin Inhibitors Based on the Structural Simplification of Sanglifehrin A.
J. Med. Chem., 60, 2017
|
|
2NXB
 
 | | Crystal structure of human Bromodomain containing protein 3 (BRD3) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, SODIUM ION | | Authors: | Filippakopoulos, P, Bullock, A, Cooper, C, Keates, K, Berridge, G, Pike, A, Bunkoczi, G, Edwards, A, Arrowsmith, C, Sundstrom, M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-17 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
2ZKO
 
 | |
1A15
 
 | | SDF-1ALPHA | | Descriptor: | STROMAL DERIVED FACTOR-1ALPHA, SULFATE ION | | Authors: | Dealwis, C.G, Fernandez, E.J, Lolis, E. | | Deposit date: | 1997-12-22 | | Release date: | 1998-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of chemically synthesized [N33A] stromal cell-derived factor 1alpha, a potent ligand for the HIV-1 "fusin" coreceptor.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3KDW
 
 | |
5RGH
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1619978933 (Mpro-x0395) | | Descriptor: | 3C-like proteinase, 5-fluoro-1-[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]-1,2,3,6-tetrahydropyridine, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
3K99
 
 | | HSP90 N-terminal domain in complex with 4-(1,3-dihydro-2H-isoindol-2-ylcarbonyl)benzene-1,3-diol | | Descriptor: | 4-(1,3-dihydro-2H-isoindol-2-ylcarbonyl)benzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Gajiwala, K.S, Davies II, J.F. | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dihydroxyphenylisoindoline amides as orally bioavailable inhibitors of the heat shock protein 90 (hsp90) molecular chaperone.
J.Med.Chem., 53, 2010
|
|
