6DIH
 
 | | Crystal structure of Tdp1 catalytic domain in complex with Sigma Aldrich compound PH004941 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxybenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-23 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
4UNO
 
 | | Crystal structure of the ETS domain of human ETV5 in complex with DNA | | Descriptor: | 5'-D(*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP)-3', 5'-D(*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP)-3', CALCIUM ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Cooper, C.D.O, Pinkas, D.M, Shrestha, L, Burgess-Brown, N, Kopec, J, Fitzpatrick, F, Tallant, C, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Gileadi, O. | | Deposit date: | 2014-05-29 | | Release date: | 2014-06-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
4V5X
 
 | | The cryo-EM structure of a 3D DNA-origami object | | Descriptor: | SCAFFOLD STRAND,SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Bai, X.C, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2012-10-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Cryo-Em Structure of a 3D DNA-Origami Object.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7M5W
 
 | | Crystal structure of the HMG-C1 domain of human capicua bound to DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*TP*TP*TP*TP*TP*CP*AP*TP*TP*CP*AP*TP*AP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*TP*AP*TP*GP*AP*AP*TP*GP*AP*AP*AP*AP*AP*GP*C)-3'), ... | | Authors: | Webb, J.P, Liew, J.J.M, Gnann, A.D, Dowling, D.P. | | Deposit date: | 2021-03-25 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular basis of DNA recognition by the HMG-box-C1 module of Capicua
Biorxiv, 2022
|
|
8RM6
 
 | | Crystal Structure of Human Androgen Receptor DNA Binding Domain Bound to its Response Element: C3(1)ARE | | Descriptor: | C3(1)ARE_Chain C, C3(1)ARE_Chain D, Isoform 2 of Androgen receptor, ... | | Authors: | Lee, X.Y, Helsen, C, Van Eynde, W, Voet, A, Claessens, F. | | Deposit date: | 2024-01-05 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural mechanism underlying variations in DNA binding by the androgen receptor.
J.Steroid Biochem.Mol.Biol., 241, 2024
|
|
6R14
 
 | | Structure of kiteplatinated dsDNA | | Descriptor: | Kiteplatin, Kiteplatinated DNA oligomer, chain A, ... | | Authors: | Margiotta, N, Papadia, P, Kubicek, K, Krejcikova, M, Gkionis, K, Sponer, J. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of kiteplatinated DNA
To Be Published
|
|
4OGC
 
 | | Crystal structure of the Type II-C Cas9 enzyme from Actinomyces naeslundii | | Descriptor: | ACETATE ION, HNH endonuclease domain protein, MAGNESIUM ION, ... | | Authors: | Jiang, F, Ma, E, Lin, S, Doudna, J.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-12 | | Last modified: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Cas9 endonucleases reveal RNA-mediated conformational activation.
Science, 343, 2014
|
|
7R08
 
 | | Abortive infection DNA polymerase Abi-P2 | | Descriptor: | Reverse transcriptase | | Authors: | Gapinska, M.A, Figiel, M, Czarnocki Cieciura, M, Nowotny, M, Zajko, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
8G8J
 
 | |
8GBF
 
 | |
6LBM
 
 | |
1VRO
 
 | | Selenium-Assisted Nucleic Acid Crystallography: Use of Phosphoroselenoates for MAD Phasing of a DNA Structure | | Descriptor: | 5'-D(*CP*(GMS)P*CP*GP*CP*G)-3', MAGNESIUM ION, SPERMINE | | Authors: | Wilds, C.J, Pattanayek, R, Pan, C, Wawrzak, Z, Egli, M. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Selenium-Assisted Nucleic Acid Crystallography: Use of Phosphoroselenoates for MAD Phasing of a DNA Structure
J.Am.Chem.Soc., 124, 2002
|
|
4U6P
 
 | |
3UN0
 
 | | Crystal Structure of MDC1 FHA Domain | | Descriptor: | Mediator of DNA damage checkpoint protein 1, SULFATE ION | | Authors: | Clapperton, J.A, Lloyd, J, Haire, L.F, Li, J, Smerdon, S.J. | | Deposit date: | 2011-11-15 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator.
Nucleic Acids Res., 40, 2012
|
|
7R7J
 
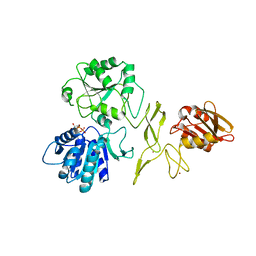 | | Crystal structure of RadD with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative DNA repair helicase RadD, ... | | Authors: | Osorio Garcia, M.A, Satyshur, K.A, Keck, J.L, Cox, M.M. | | Deposit date: | 2021-06-24 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | X-ray crystal structure of the Escherichia coli RadD DNA repair protein bound to ADP reveals a novel zinc ribbon domain.
Plos One, 17, 2022
|
|
1I0T
 
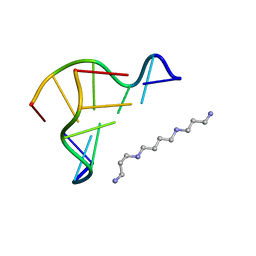 | | 0.6 A STRUCTURE OF Z-DNA CGCGCG | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', SPERMINE | | Authors: | Tereshko, V, Wilds, C.J, Minasov, G, Prakash, T.P, Maier, M.A, Howard, A, Wawrzak, Z, Manoharan, M, Egli, M. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.6 Å) | | Cite: | Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments.
Nucleic Acids Res., 29, 2001
|
|
7OPD
 
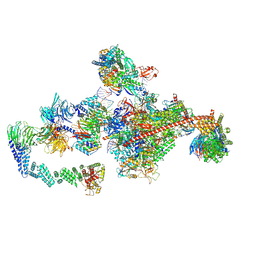 | | Pol II-CSB-CRL4CSA-UVSSA-SPT6-PAF (Structure 5) | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-31 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OPC
 
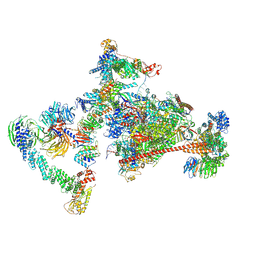 | | Pol II-CSB-CRL4CSA-UVSSA-SPT6-PAF (Structure 4) | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-31 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
6R6D
 
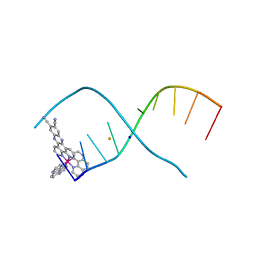 | | [Ru(TAP)2(11,12-CN2-dppz)]2+ bound to d(TCGGCGCCGA)2 | | Descriptor: | BARIUM ION, DNA (5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3'), Ruthenium (bis-(tetraazaphenanthrene)) (11,12-dicyano-dipyridophenazine), ... | | Authors: | McQuaid, K.T, Hall, J.P, Cardin, C.J. | | Deposit date: | 2019-03-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | X-ray Crystal Structures Show DNA Stacking Advantage of Terminal Nitrile Substitution in Ru-dppz Complexes.
Chemistry, 24, 2018
|
|
7R8I
 
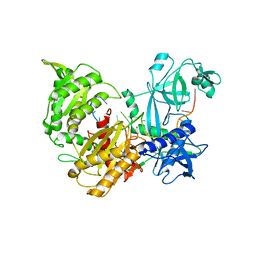 | |
7OO3
 
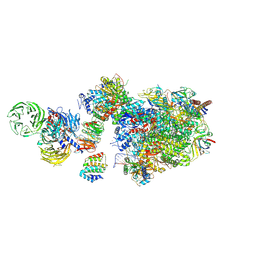 | | Pol II-CSB-CSA-DDB1-UVSSA (Structure1) | | Descriptor: | CSB element, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-26 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OOB
 
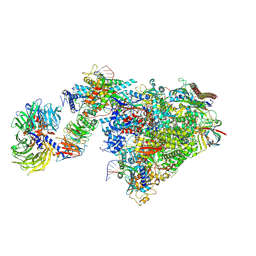 | | Pol II-CSB-CSA-DDB1-UVSSA-ADPBeF3 (Structure2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA damage-binding protein 1, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7O4I
 
 | | Yeast RNA polymerase II transcription pre-initiation complex with initial transcription bubble | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | Deposit date: | 2021-04-06 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
7O4J
 
 | | Yeast RNA polymerase II transcription pre-initiation complex (consensus) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | Deposit date: | 2021-04-06 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
8SKI
 
 | |
