1C0Y
 
 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED OPPOSITE DA AT A TEMPLATE-PRIMER JUNCTION | | Descriptor: | 2-AMINOFLUORENE, DNA (5'-D(*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*GP*TP*AP*GP*C)-3') | | Authors: | Gu, Z, Gorin, A, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1999-07-19 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of aminofluorene [AF]-stacked conformers of the syn [AF]-C8-dG adduct positioned opposite dC or dA at a template-primer junction.
Biochemistry, 38, 1999
|
|
1EZ1
 
 | | STRUCTURE OF ESCHERICHIA COLI PURT-ENCODED GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE COMPLEXED WITH MG, AMPPNP, AND GAR | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ACETATE ION, GLYCINAMIDE RIBONUCLEOTIDE, ... | | Authors: | Thoden, J.B, Firestine, S, Nixon, A, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2000-05-09 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular structure of Escherichia coli PurT-encoded glycinamide ribonucleotide transformylase.
Biochemistry, 39, 2000
|
|
1INF
 
 | |
6DFG
 
 | | BG505 MD39 SOSIP trimer in complex with mature BG18 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Ozorowski, G, Steichen, J.M, Schief, W.R, Ward, A.B. | | Deposit date: | 2018-05-14 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
100D
 
 | |
169D
 
 | | THE SOLUTION STRUCTURE OF THE R(GCG)D(TATACCC):D(GGGTATACGC) OKAZAKI FRAGMENT CONTAINS TWO DISTINCT DUPLEX MORPHOLOGIES CONNECTED BY A JUNCTION | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*AP*TP*AP*CP*GP*C)-3'), DNA/RNA (5'-R(*GP*CP*G)-D(P*TP*AP*TP*AP*CP*CP*C)-3') | | Authors: | Salazar, M, Fedoroff, O.Y, Zhu, L, Reid, B.R. | | Deposit date: | 1994-04-11 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the r(gcg)d(TATACCC):d(GGGTATACGC) Okazaki fragment contains two distinct duplex morphologies connected by a junction.
J.Mol.Biol., 241, 1994
|
|
3RWD
 
 | | rhesus macaque MHC class I molecule Mamu-B*17-IW11 | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I, Nef IW11 peptide from Protein Nef | | Authors: | Wu, Y, Gao, F, Liu, J, Qi, J.X, Price, D.A, Gao, G.F. | | Deposit date: | 2011-05-09 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structural basis of diverse peptide accommodation by the rhesus macaque MHC class I molecule Mamu-B*17: insights into immune protection from simian immunodeficiency virus
J.Immunol., 187, 2011
|
|
132D
 
 | |
137D
 
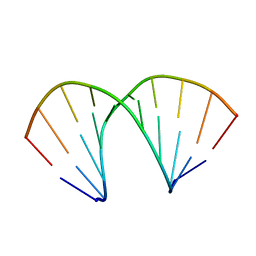 | | A-DNA DECAMER D(GCGGGCCCGC)-ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | DNA (5'-D(*GP*CP*GP*GP*GP*CP*CP*CP*GP*C)-3') | | Authors: | Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures.
Biochemistry, 32, 1993
|
|
5B8I
 
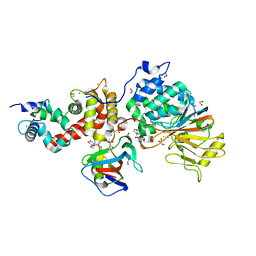 | | Crystal structure of Calcineurin A and Calcineurin B in complex with FKBP12 and FK506 from Coccidioides immitis RS | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fox III, D, Dranow, D.M, Lorimer, D.D, Edwards, T.E. | | Deposit date: | 2015-05-03 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents.
Nat Commun, 10, 2019
|
|
198D
 
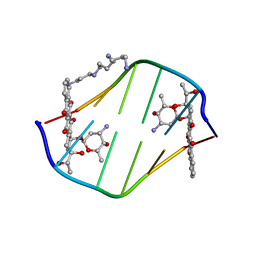 | | A TRIGONAL FORM OF THE IDARUBICIN-D(CGATCG) COMPLEX: CRYSTAL AND MOLECULAR STRUCTURE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), IDARUBICIN, SPERMINE | | Authors: | Dautant, A, Langlois D'Estaintot, B, Gallois, B, Brown, T, Hunter, W.N. | | Deposit date: | 1994-11-28 | | Release date: | 1995-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A trigonal form of the idarubicin:d(CGATCG) complex; crystal and molecular structure at 2.0 A resolution.
Nucleic Acids Res., 23, 1995
|
|
3FPX
 
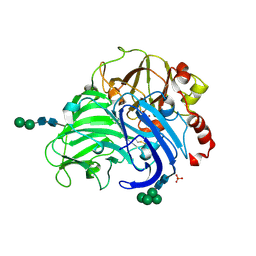 | | Native fungus laccase from Trametes hirsuta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Fedorova, T.V, Stepanova, E.V, Cherkashin, E.A, Kurzeev, S.A, Strokopytov, B.V, Lamzin, V.S, Koroleva, O.V. | | Deposit date: | 2009-01-06 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of native laccase from Coriolus hirsutus at 1.8 A resolution
To be Published
|
|
1NUJ
 
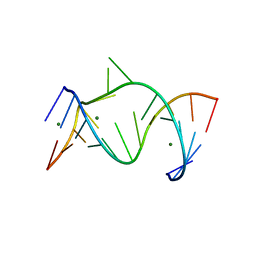 | | THE LEADZYME STRUCTURE BOUND TO MG(H20)6(II) AT 1.8 A RESOLUTION | | Descriptor: | 5'-R(*CP*GP*GP*AP*CP*CP*GP*AP*GP*CP*CP*AP*G)-3', 5'-R(*GP*CP*UP*GP*GP*GP*AP*GP*UP*CP*C)-3', MAGNESIUM ION | | Authors: | Wedekind, J.E, Mckay, D.B. | | Deposit date: | 2003-01-31 | | Release date: | 2003-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the leadzyme at 1.8 A resolution: metal ion binding and the implications
for catalytic mechanism and allo site ion regulation.
BIOCHEMISTRY, 42, 2003
|
|
3RO5
 
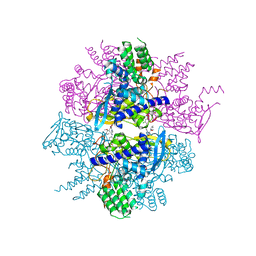 | | Crystal structure of influenza A virus nucleoprotein with ligand | | Descriptor: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Pearce, B.C, Edavettal, S, McDonnell, P.A, Lewis, H.A, Steinbacher, S, Baldwin, E.T, Langley, D.R, Maskos, K, Mortl, M, Kiefersauer, R. | | Deposit date: | 2011-04-25 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Inhibition of influenza virus replication via small molecules that induce the formation of higher-order nucleoprotein oligomers.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1GQG
 
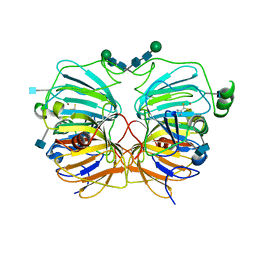 | | Quercetin 2,3-dioxygenase in complex with the inhibitor diethyldithiocarbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2001-11-23 | | Release date: | 2002-06-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Analysis of the Copper-Dependent Quercetin 2,3-Dioxygenase.1.Ligand-Induced Coordination Changes Probed by X-Ray Crystallography: Inhibition, Ordering Effect and Mechanistic Insights
Biochemistry, 41, 2002
|
|
1MCE
 
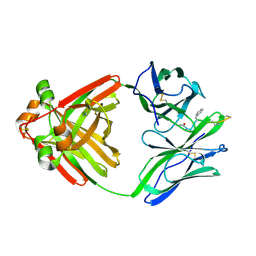 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | Immunoglobulin lambda-1 light chain, PEPTIDE N-ACETYL-L-GLN-D-PHE-L-HIS-D-PRO-B-ALA-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2022-04-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
3RWC
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*17-IW9 | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I, Nef IW9 peptide from Protein Nef | | Authors: | Wu, Y, Gao, F, Liu, J, Qi, J.X, Price, D.A, Gao, G.F. | | Deposit date: | 2011-05-09 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural basis of diverse peptide accommodation by the rhesus macaque MHC class I molecule Mamu-B*17: insights into immune protection from simian immunodeficiency virus
J.Immunol., 187, 2011
|
|
3JTG
 
 | | Crystal structure of mouse Elf3 C-terminal DNA-binding domain in complex with type II TGF-beta receptor promoter DNA | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*CP*AP*GP*GP*AP*AP*AP*CP*TP*CP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*GP*TP*TP*TP*CP*CP*TP*GP*TP*TP*T)-3'), ETS-related transcription factor Elf-3 | | Authors: | Tahirov, T.H, Babayeva, N.D, Agarkar, V.B, Rizzino, A. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of mouse Elf3 C-terminal DNA-binding domain in complex with type II TGF-beta receptor promoter DNA.
J.Mol.Biol., 397, 2010
|
|
3RWE
 
 | | rhesus macaque MHC class I molecule Mamu-B*17-FW9 | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, Major histocompatibility complex class I, ... | | Authors: | Wu, Y, Gao, F, Liu, J, Qi, J.X, Price, D.A, Gao, G.F. | | Deposit date: | 2011-05-09 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of diverse peptide accommodation by the rhesus macaque MHC class I molecule Mamu-B*17: insights into immune protection from simian immunodeficiency virus
J.Immunol., 187, 2011
|
|
1LNE
 
 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CADMIUM ION, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
1HEZ
 
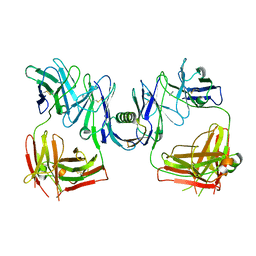 | | Structure of P. magnus protein L bound to a human IgM Fab. | | Descriptor: | HEAVY CHAIN OF IG, IMIDAZOLE, KAPPA LIGHT CHAIN OF IG, ... | | Authors: | Graille, M, Stura, E.A, Housden, N.G, Bottomley, S.P, Taussig, M.J, Sutton, B.J, Gore, M.G, Charbonnier, J.B. | | Deposit date: | 2000-11-27 | | Release date: | 2001-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex between Peptostreptococcus Magnus Protein L and a Human Antibody Reveals Structural Convergence in the Interaction Modes of Fab Binding Modes
Structure, 9, 2001
|
|
1LE5
 
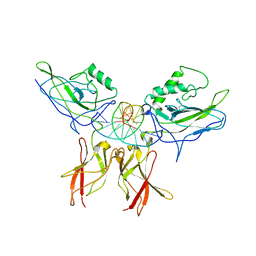 | | Crystal structure of a NF-kB heterodimer bound to an IFNb-kB | | Descriptor: | 5'-D(*AP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3', 5'-D(*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*T)-3', Nuclear factor NF-kappa-B p50 subunit, ... | | Authors: | Berkowitz, B, Huang, D.B, Chen-Park, F.E, Sigler, P.B, Ghosh, G. | | Deposit date: | 2002-04-09 | | Release date: | 2003-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The X-ray crystal structure of the
NF-kB p50/p65 heterodimer bound
to the Interferon beta-kB site
J.Biol.Chem., 277, 2002
|
|
3BC9
 
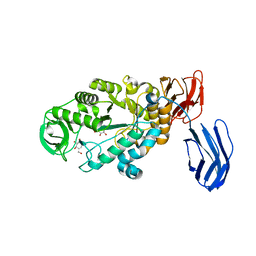 | | Alpha-amylase B in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
1FLC
 
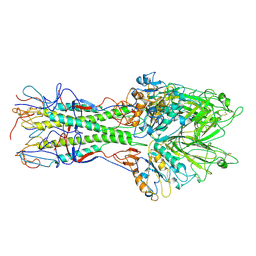 | | X-RAY STRUCTURE OF THE HAEMAGGLUTININ-ESTERASE-FUSION GLYCOPROTEIN OF INFLUENZA C VIRUS | | Descriptor: | HAEMAGGLUTININ-ESTERASE-FUSION GLYCOPROTEIN, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rosenthal, P.B, Zhang, X, Formanowski, F, Fitz, W, Wong, C.H, Meier-Ewert, H, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1999-02-22 | | Release date: | 2000-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus.
Nature, 396, 1998
|
|
3OL2
 
 | | Receptor-ligand structure of Human Semaphorin 4D with Plexin B1. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-B1, ... | | Authors: | Janssen, B.J.C, Robinson, R.A, Perez-Branguli, F, Bell, C.H, Mitchell, C.J, Siebold, C, Jones, E.Y. | | Deposit date: | 2010-08-25 | | Release date: | 2010-10-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis of semaphorin-plexin signalling.
Nature, 467, 2010
|
|
