5RWL
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z1348371854 | | Descriptor: | 5-(1,4-oxazepan-4-yl)pyridine-2-carbonitrile, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
1NK8
 
 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER A SINGLE ROUND OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP. | | Descriptor: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | Authors: | Johnson, S.J, Beese, L.S. | | Deposit date: | 2003-01-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
5RZ1
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z27682767 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
7JXI
 
 | | EGFR kinase (T790M/V948R) in complex with PF-06747775 | | Descriptor: | Epidermal growth factor receptor, N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-methyl-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors
Nat Commun, 13, 2022
|
|
1NJZ
 
 | |
1NK9
 
 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER TWO ROUNDS OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP AND DGTP. | | Descriptor: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | Authors: | Johnson, S.J, Beese, L.S. | | Deposit date: | 2003-01-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
3GK1
 
 | | X-ray structure of bovine SBi132,Ca(2+)-S100B | | Descriptor: | 2-[(5-hex-1-yn-1-ylfuran-2-yl)carbonyl]-N-methylhydrazinecarbothioamide, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
5RWR
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2856434783 | | Descriptor: | DIMETHYL SULFOXIDE, N-benzyl-1-(4-fluorophenyl)methanamine, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5TOW
 
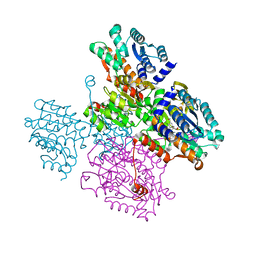 | | Crystal structure of the inactive form of S-adenosyl-L-homocysteine hydrolase from Thermotoga maritima in ternary complex with NADH and Adenosine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE, ... | | Authors: | Czyrko, J, Brzezinski, K. | | Deposit date: | 2016-10-19 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | S-adenosyl-L-homocysteine hydrolase from a hyperthermophile (Thermotoga maritima) is expressed in Escherichia coli in inactive form - Biochemical and structural studies.
Int. J. Biol. Macromol., 104, 2017
|
|
5RYJ
 
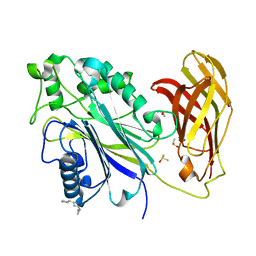 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2177153697 | | Descriptor: | 3-cyclopropyl-5-[(3R)-pyrrolidin-3-yl]-1,2,4-oxadiazole, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
4U6W
 
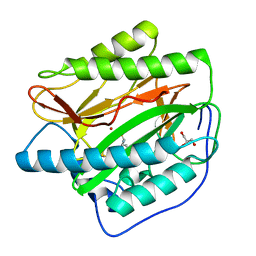 | |
5RZQ
 
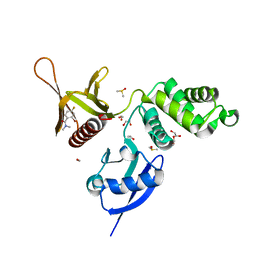 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z100642432 | | Descriptor: | 1,2-ETHANEDIOL, 2-bromo-4-fluoro-N,N-dimethylbenzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
3QMA
 
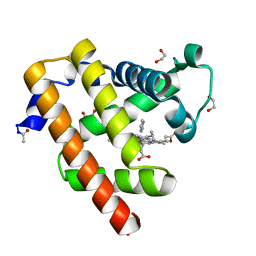 | | Blackfin tuna myoglobin imidazole complex, atomic resolution | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Myoglobin, ... | | Authors: | de Serrano, V.S, Rodriguez, M.M, Schreiter, E.R. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | High Resolution Structures of Blackfin Tuna Myoglobin
To be Published
|
|
7K0U
 
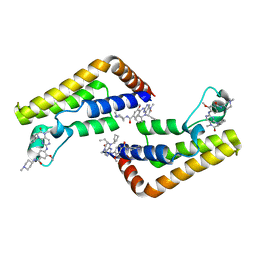 | |
5REA
 
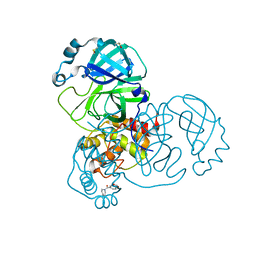 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z31432226 | | Descriptor: | (azepan-1-yl)(2H-1,3-benzodioxol-5-yl)methanone, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
1X74
 
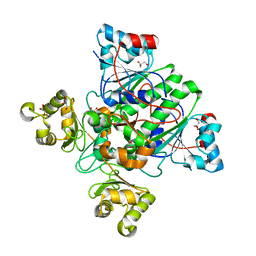 | | Alpha-methylacyl-CoA racemase from Mycobacterium tuberculosis- mutational and structural characterization of the fold and active site | | Descriptor: | 2-methylacyl-CoA racemase, GLYCEROL, PHOSPHATE ION | | Authors: | Kalle, S, Bhaumik, P, Schmitz, W, Kotti, T.J, Conzelmann, E, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2004-08-13 | | Release date: | 2005-01-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | {alpha}-Methylacyl-CoA Racemase from Mycobacterium tuberculosis: MUTATIONAL AND STRUCTURAL CHARACTERIZATION OF THE ACTIVE SITE AND THE FOLD
J.Biol.Chem., 280, 2005
|
|
3QT8
 
 | | Crystal structure of mutant S192A Staphylococcus epidermidis mevalonate diphosphate decarboxylase complexed with inhibitor 6-FMVAPP | | Descriptor: | (3R)-3-(fluoromethyl)-3-hydroxy-5-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}pentanoic acid, GLYCEROL, Mevalonate diphosphate decarboxylase | | Authors: | Barta, M.L, Skaff, A.D, McWhorter, W.J, Miziorko, H.M, Geisbrecht, B.V. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of Staphylococcus epidermidis mevalonate diphosphate decarboxylase bound to inhibitory analogs reveal new insight into substrate binding and catalysis.
J.Biol.Chem., 286, 2011
|
|
5R99
 
 | |
4D2J
 
 | | Crystal structure of F16BP Aldolase from Toxoplasma gondii (TgALD1) | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, FRUCTOSE-BISPHOSPHATE ALDOLASE, ... | | Authors: | Tonkin, M.L, Ramaswamy, R, Halavaty, A.S, Ruan, J, Igarashi, M, Ngo, H.M, Boulanger, M.J. | | Deposit date: | 2014-05-09 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Functional Divergence of the Aldolase Fold in Toxoplasma Gondii.
J.Mol.Biol., 427, 2015
|
|
1NHY
 
 | | Crystal Structure of the GST-like Domain of Elongation Factor 1-gamma from Saccharomyces cerevisiae. | | Descriptor: | Elongation factor 1-gamma 1, SULFATE ION | | Authors: | Jeppesen, M.G, Ortiz, P, Kinzy, T.G, Andersen, G.R, Nyborg, J. | | Deposit date: | 2002-12-20 | | Release date: | 2003-01-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of the Glutathione S-Transferase-like Domain of Elongation Factor 1B{gamma} from Saccharomyces cerevisiae.
J.Biol.Chem., 278, 2003
|
|
2CF9
 
 | | Complex of recombinant human thrombin with an inhibitor | | Descriptor: | 4-[(1R,3AS,4R,8AS,8BR)-1-ISOPROPYL-2-(4-METHOXYBENZYL)-3-OXODECAHYDROPYRROLO[3,4-A]PYRROLIZIN-4-YL]BENZENECARBOXIMIDAMIDE, CALCIUM ION, HIRUDIN IIIA, ... | | Authors: | Schweizer, E, Hoffmann-Roeder, A, Olsen, J.A, Obst-Sander, U, Wagner, B, Kansy, M, Banner, D.W, Diederich, F. | | Deposit date: | 2006-02-17 | | Release date: | 2006-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Multipolar Interactions in the D Pocket of Thrombin: Large Differences between Tricyclic Imide and Lactam Inhibitors.
Org.Biomol.Chem., 4, 2006
|
|
4TUY
 
 | | Tubulin-Rhizoxin complex | | Descriptor: | (1R,2R,3E,5R,7R,8S,10S,13E,16R)-8-hydroxy-10-[(2S,3R,4E,6E,8E)-3-methoxy-4,8-dimethyl-9-(2-methyl-1,3-oxazol-4-yl)nona-4,6,8-trien-2-yl]-2,7-dimethyl-6,11,19-trioxatricyclo[14.3.1.0~5,7~]icosa-3,13-diene-12,18-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Bargsten, K, Diaz, J.F, Marsh, M, Cuevas, C, Liniger, M, Neuhaus, C, Andreu, J.M, Altmann, K.H, Steinmetz, M.O. | | Deposit date: | 2014-06-25 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4IXO
 
 | |
6BXA
 
 | | Crystal structure of N-terminal fragment of Zebrafish Toll-Like Receptor 5 (TLR5) with Lamprey Variable Lymphocyte Receptor 2 (VLR2) bound | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gunn, R.J, Wilson, I.A, Cooper, M.D, Herrin, B.R. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | VLR Recognition of TLR5 Expands the Molecular Characterization of Protein Antigen Binding by Non-Ig-based Antibodies.
J. Mol. Biol., 430, 2018
|
|
1NJW
 
 | |
