5V54
 
 | | Crystal structure of 5-HT1B receptor in complex with methiothepin | | Descriptor: | 1-methyl-4-[(5~{S})-3-methylsulfanyl-5,6-dihydrobenzo[b][1]benzothiepin-5-yl]piperazine, 5-hydroxytryptamine receptor 1B,OB-1 fused 5-HT1b receptor,5-hydroxytryptamine receptor 1B | | Authors: | Yin, W.C, Zhou, X.E, Yang, D, de Waal, P, Wang, M.T, Dai, A, Cai, X, Huang, C.Y, Liu, P, Yin, Y, Liu, B, Caffrey, M, Melcher, K, Xu, Y, Wang, M.W, Xu, H.E, Jiang, Y. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | A common antagonistic mechanism for class A GPCRs revealed by the structure of the human 5-HT1B serotonin receptor bound to an antagonist
Cell Discov, 2018
|
|
7GU1
 
 | |
5RZ3
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z1259341037 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
7GUB
 
 | |
7GUA
 
 | |
7GUC
 
 | |
7GTJ
 
 | |
7GSV
 
 | |
4BUW
 
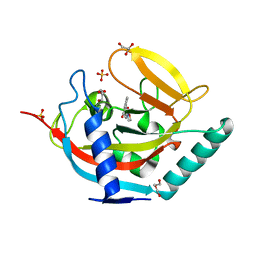 | | Crystal structure of human tankyrase 2 in complex with 2-(4-(2-oxo-1, 3-oxazolidin-3-yl)phenyl)-3,4-dihydroquinazolin-4-one | | Descriptor: | 2-[4-(2-OXO-1,3-OXAZOLIDIN-3-YL)PHENYL]-3,4-DIHYDROQUINAZOLIN-4-ONE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
5BXL
 
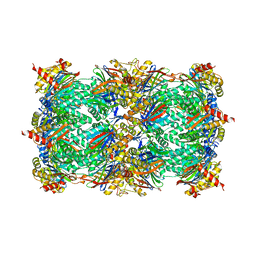 | | Yeast 20S proteasome beta2-G170A mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-06-09 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defective immuno- and thymoproteasome assembly causes severe immunodeficiency.
Sci Rep, 8, 2018
|
|
5RZY
 
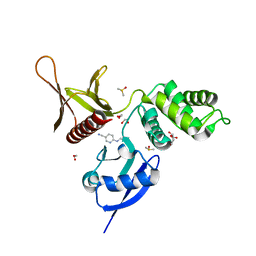 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z219104216 | | Descriptor: | 1,2-ETHANEDIOL, 6-(ethylamino)pyridine-3-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
7GTV
 
 | |
3IU2
 
 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90096 | | Descriptor: | (2R)-2-{4-hydroxy-5-methoxy-2-[3-(4-methylpiperazin-1-yl)propyl]phenyl}-3-pyridin-3-yl-1,3-thiazolidin-4-one, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-29 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90096
To be Published
|
|
7GT1
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOMB000209a | | Descriptor: | (2S)-2-(2-chloro-6-fluorophenyl)-2,3-dihydroquinazolin-4(1H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
3R2U
 
 | | 2.1 Angstrom Resolution Crystal Structure of Metallo-beta-lactamase from Staphylococcus aureus subsp. aureus COL | | Descriptor: | CHLORIDE ION, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-14 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of Metallo-beta-lactamase Family Protein from Staphylococcus aureus subsp. aureus COL
TO BE PUBLISHED
|
|
3NDY
 
 | |
1IRV
 
 | |
4R8Y
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((R)-1-(2-cyclopentylacetyl)pyrrolidin-3-yl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-{[(3R)-1-(cyclopentylacetyl)pyrrolidin-3-yl]methyl}-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Caldwell, J.P, Strickland, C. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2ZLY
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
2ZM9
 
 | | Structure of 6-Aminohexanoate-dimer Hydrolase, A61V/S112A/A124V/R187S/F264C/G291R/G338A/D370Y mutant (Hyb-S4M94) with Substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
1IOP
 
 | | INCORPORATION OF A HEMIN WITH THE SHORTEST ACID SIDE-CHAINS INTO MYOGLOBIN | | Descriptor: | 6,7-DICARBOXYL-1,2,3,4,5,8-HEXAMETHYLHEMIN, CYANIDE ION, MYOGLOBIN, ... | | Authors: | Igarashi, N, Neya, S, Funasaki, N, Tanaka, N. | | Deposit date: | 1997-12-12 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of 6,7-dicarboxyheme-substituted myoglobin
Biochemistry, 37, 1998
|
|
5RXG
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2856434909 | | Descriptor: | (2S)-1-{[(2,6-dichlorophenyl)methyl]amino}propan-2-ol, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RXV
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z45527714 | | Descriptor: | 2-chlorobenzene-1-thiol, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RHA
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z147647874 (Mpro-x2779) | | Descriptor: | 1-{4-[(thiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5E9V
 
 | | Crystal structure of BRD9 bromodomain in complex with an indolizine ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-[1-(imidazo[1,2-a]pyridin-5-yl)-7-(morpholin-4-yl)indolizin-3-yl]ethanone, Bromodomain-containing protein 9 | | Authors: | Tallant, C, Hay, D.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Schofield, C.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-10-15 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of BRD9 bromodomain in complex with an indolizine ligand
To Be Published
|
|
