1HI3
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine 2'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
2V4E
 
 | | A non-cytotoxic DsRed variant for whole-cell labeling | | Descriptor: | RED FLUORESCENT PROTEIN DRFP583 | | Authors: | Strack, R.L, Strongin, D.E, Bhattacharyya, D, Tao, W, Berman, A, Broxmeyer, H.E, Keenan, R.J, Glick, B.S. | | Deposit date: | 2008-09-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Noncytotoxic Dsred Variant for Whole-Cell Labeling.
Nat.Methods, 5, 2008
|
|
2VFZ
 
 | | CRYSTAL STRUCTURE OF ALPHA-1,3 GALACTOSYLTRANSFERASE (R365K) IN COMPLEX WITH UDP-2F-GALACTOSE | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYL TRANSFERASE, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUOROGALACTOSE | | Authors: | Jamaluddin, H, Tumbale, P, Withers, S.G, Acharya, K.R, Brew, K. | | Deposit date: | 2007-11-06 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes Induced by Binding Udp-2F-Galactose to Alpha-1,3 Galactosyltransferase-Implications for Catalysis
J.Mol.Biol., 369, 2007
|
|
2VAH
 
 | | Solution structure of a B-DNA hairpin at low pressure. | | Descriptor: | 5'-D(*AP*GP*GP*AP*TP*CP*CP*TP*UP*TP *TP*GP*GP*AP*TP*CP*CP*T)-3' | | Authors: | Williamson, M.P, Wilton, D.J, Ghosh, M, Chary, K.V.A, Akasaka, K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Change in a B-DNA Helix with Hydrostatic Pressure
Nucleic Acids Res., 36, 2008
|
|
2XSV
 
 | | Crystal structure of L69A mutant Acinetobacter radioresistens catechol 1,2 dioxygenase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, CATECHOL 1,2 DIOXYGENASE, FE (III) ION | | Authors: | Micalella, C, Martignon, S, Bruno, S, Rizzi, M. | | Deposit date: | 2010-09-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Crystallography, Mass Spectrometry and Single Crystal Microspectrophotometry: A Multidisciplinary Characterization of Catechol 1,2 Dioxygenase.
Biochim.Biophys.Acta, 1814, 2011
|
|
2XW7
 
 | | Structure of Mycobacterium smegmatis putative reductase MS0308 | | Descriptor: | DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Evangelopoulos, D, Gupta, A, Lack, N, Cronin, N, Daviter, T, Sim, E, Keep, N.H, Bhakta, S. | | Deposit date: | 2010-11-01 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of an Oxidoreductase from the Arylamine N-Acetyltransferase Operon in Mycobacterium Smegmatis.
FEBS J., 278, 2011
|
|
2FIB
 
 | | RECOMBINANT HUMAN GAMMA-FIBRINOGEN CARBOXYL TERMINAL FRAGMENT (RESIDUES 143-411) COMPLEXED TO THE PEPTIDE GLY-PRO-ARG-PRO AT PH 6.0 | | Descriptor: | CALCIUM ION, FIBRINOGEN, GLY-PRO-ARG-PRO | | Authors: | Pratt, K.P, Cote, H.C.F, Chung, D.W, Stenkamp, R.E, Davie, E.W. | | Deposit date: | 1997-06-03 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The primary fibrin polymerization pocket: three-dimensional structure of a 30-kDa C-terminal gamma chain fragment complexed with the peptide Gly-Pro-Arg-Pro.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
3C9J
 
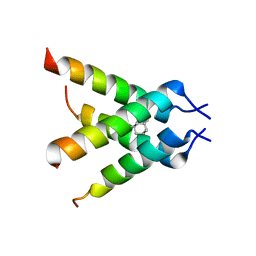 | | The Crystal structure of Transmembrane domain of M2 protein and Amantadine complex | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, Proton Channel protein M2, transmembrane segment | | Authors: | Stouffer, A.L, Acharya, R, Salom, D. | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the function and inhibition of an influenza virus proton channel
Nature, 451, 2008
|
|
1LUL
 
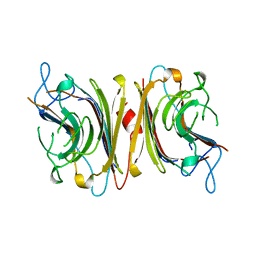 | | DB58, A LEGUME LECTIN FROM DOLICHOS BIFLORUS | | Descriptor: | CALCIUM ION, LECTIN DB58, MANGANESE (II) ION | | Authors: | Hamelryck, T.W, Bouckaert, J, Dao-Thi, M.H, Wyns, L, Etzler, M, Loris, R. | | Deposit date: | 1998-06-30 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
2JWM
 
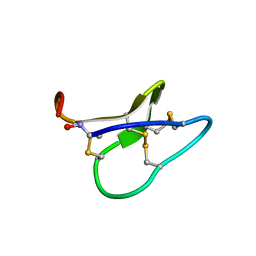 | |
2YDM
 
 | | Structural characterization of angiotensin-I converting enzyme in complex with a selenium analogue of captopril | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN CONVERTING ENZYME, CHLORIDE ION, ... | | Authors: | Akif, M, Masuyer, G, Schwager, S.L.U, Bhuyan, B.J, Mugesh, G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2011-03-22 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural Characterization of Angiotensin-I Converting Enzyme in Complex with a Selenium Analogue of Captopril
FEBS J., 278, 2011
|
|
2YQU
 
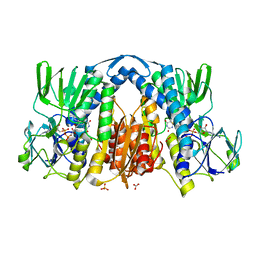 | | Crystal structures and evolutionary relationship of two different lipoamide dehydrogenase(E3s) from Thermus thermophilus | | Descriptor: | 2-oxoglutarate dehydrogenase E3 component, CARBONATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kondo, H, Hossain, M.T, Adachi, W, Nakai, T, Kamiya, N, Kuramitsu, K. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures and evolutionary relationship of two different lipoamide dehydrogenase(E3s) from Thermus thermophilus
To be Published
|
|
2QGD
 
 | |
390D
 
 | | STRUCTURAL VARIABILITY AND NEW INTERMOLECULAR INTERACTIONS OF Z-DNA IN CRYSTALS OF D(PCPGPCPGPCPG) | | Descriptor: | DNA (5'-D(P*CP*GP*CP*GP*CP*G)-3') | | Authors: | Malinina, L, Tereshko, V, Ivanova, E, Subirana, J.A, Zarytova, V, Nekrasov, Y. | | Deposit date: | 1998-04-20 | | Release date: | 1998-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural variability and new intermolecular interactions of Z-DNA in crystals of d(pCpGpCpGpCpG).
Biophys.J., 74, 1998
|
|
2XQT
 
 | | Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases | | Descriptor: | ATP SYNTHASE C CHAIN, CYMAL-4, DICYCLOHEXYLUREA | | Authors: | Pogoryelov, D, Krah, A, Langer, J, Yildiz, O, Faraldo-Gomez, J.D, Meier, T. | | Deposit date: | 2010-09-07 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Microscopic Rotary Mechanism of Ion Translocation in the Fo Complex of ATP Synthases
Nat.Chem.Biol., 6, 2010
|
|
2XQS
 
 | | Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases | | Descriptor: | ATP SYNTHASE C CHAIN, CYMAL-4 | | Authors: | Pogoryelov, D, Krah, A, Langer, J, Yildiz, O, Faraldo-Gomez, J.D, Meier, T. | | Deposit date: | 2010-09-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Microscopic Rotary Mechanism of Ion Translocation in the Fo Complex of ATP Synthases
Nat.Chem.Biol., 6, 2010
|
|
2QGB
 
 | | Human transthyretin (TTR) in Apo-form | | Descriptor: | Transthyretin | | Authors: | Connelly, S, Wilson, I.A. | | Deposit date: | 2007-06-28 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and structural evaluation of highly selective 2-arylbenzoxazole-based transthyretin amyloidogenesis inhibitors.
J.Med.Chem., 51, 2008
|
|
3F39
 
 | | Apoferritin: complex with phenol | | Descriptor: | CADMIUM ION, Ferritin light chain, PHENOL, ... | | Authors: | Vedula, L.S, Economou, N.J, Rossi, M.J, Eckenhoff, R.G, Loll, P.J. | | Deposit date: | 2008-10-30 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A unitary anesthetic binding site at high resolution.
J.Biol.Chem., 284, 2009
|
|
3F36
 
 | | Apoferritin: complex with 2-isopropylphenol | | Descriptor: | 2-(1-methylethyl)phenol, CADMIUM ION, Ferritin light chain, ... | | Authors: | Vedula, L.S, Economou, N.J, Rossi, M.J, Eckenhoff, R.G, Loll, P.J. | | Deposit date: | 2008-10-30 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A unitary anesthetic binding site at high resolution.
J.Biol.Chem., 284, 2009
|
|
1G6D
 
 | | STRUCTURE OF PEPTIDYL-D(CGCAATTGCG) IN THE PRESENCE OF ZINC IONS | | Descriptor: | PEPTIDYL-D(CGCAATTGCG), ZINC ION | | Authors: | Soler-Lopez, M, Malinina, L, Tereshko, V, Zarytova, V, Subirana, J.A. | | Deposit date: | 2000-11-04 | | Release date: | 2002-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interaction of zinc ions with d(CGCAATTGCG) in a 2.9 A resolution X-ray structure.
J.Biol.Inorg.Chem., 7, 2002
|
|
2YFE
 
 | |
2PFR
 
 | | Human N-acetyltransferase 2 | | Descriptor: | Arylamine N-acetyltransferase 2, COENZYME A, SULFATE ION, ... | | Authors: | Tempel, W, Wu, H, Dombrovski, L, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Grant, D.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The Crystal Structure of Human N-acetyltransferase 2 in complex with CoA
to be published
|
|
2YGM
 
 | | THE X-RAY CRYSTAL STRUCTURE OF TANDEM CBM51 MODULES OF SP3GH98, THE FAMILY 98 GLYCOSIDE HYDROLASE FROM STREPTOCOCCUS PNEUMONIAE SP3-BS71, IN COMPLEX WITH THE BLOOD GROUP B ANTIGEN | | Descriptor: | BLOOD GROUP A-AND B-CLEAVING ENDO-BETA-GALACTOSIDASE, CALCIUM ION, SODIUM ION, ... | | Authors: | Higgins, M.A, Ficko-Blean, E, Wright, C, Meloncelli, P.J, Lowary, T.L, Boraston, A.B. | | Deposit date: | 2011-04-19 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Overall Architecture and Receptor Binding of Pneumococcal Carbohydrate Antigen Hydrolyzing Enzymes.
J.Mol.Biol., 411, 2011
|
|
1BJT
 
 | | TOPOISOMERASE II RESIDUES 409-1201 | | Descriptor: | TOPOISOMERASE II | | Authors: | Fass, D, Bogden, C.E, Berger, J.M. | | Deposit date: | 1998-06-29 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands.
Nat.Struct.Biol., 6, 1999
|
|
2YNJ
 
 | | GroEL at sub-nanometer resolution by Constrained Single Particle Tomography | | Descriptor: | 60 KDA CHAPERONIN, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Bartesaghi, A, Lecumberry, F, Sapiro, G, Subramaniam, S. | | Deposit date: | 2012-10-15 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Protein Secondary Structure Determination by Constrained Single-Particle Cryo-Electron Tomography
Structure, 20, 2012
|
|
