5W7V
 
 | |
5T86
 
 | | Crystal structure of CDI complex from E. coli A0 34/86 | | Descriptor: | ACETATE ION, CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Stols, L, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of CDI complex from E. coli A0 34/86
To Be Published
|
|
5TJ2
 
 | |
6N3B
 
 | | SegA-asym, conformation of TDP-43 low complexity domain segment A asym | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-11-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of four polymorphic TDP-43 amyloid cores.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5T8Q
 
 | |
8XH1
 
 | |
8XH0
 
 | |
8XH2
 
 | |
6HGF
 
 | | Crystal structure of Alpha1-antichymotrypsin variant NewBG-II: a new binding globulin in complex with cortisol | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, 1,2-ETHANEDIOL, Alpha-1-antichymotrypsin | | Authors: | Schmidt, K, Muller, Y.A. | | Deposit date: | 2018-08-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | NewBG: A surrogate corticosteroid-binding globulin with an unprecedentedly high ligand release efficacy.
J.Struct.Biol., 207, 2019
|
|
6HGG
 
 | | Crystal structure of Alpha1-antichymotrypsin variant NewBG-III: a new binding globulin in complex with cortisol | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, 1,2-ETHANEDIOL, Alpha-1-antichymotrypsin | | Authors: | Schmidt, K, Muller, Y.A. | | Deposit date: | 2018-08-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | NewBG: A surrogate corticosteroid-binding globulin with an unprecedentedly high ligand release efficacy.
J.Struct.Biol., 207, 2019
|
|
6HGM
 
 | |
5TJ3
 
 | | Crystal structure of wild type alkaline phosphatase PafA to 1.7A resolution | | Descriptor: | Alkaline phosphatase PafA, ZINC ION | | Authors: | Lyubimov, A.Y, Sunden, F, Ressl, S, Herschlag, D. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic and Evolutionary Insights from Comparative Enzymology of Phosphomonoesterases and Phosphodiesterases across the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 138, 2016
|
|
6ARC
 
 | |
5T8O
 
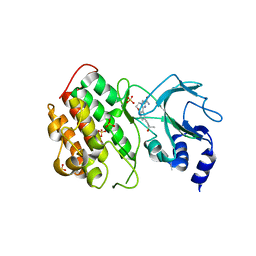 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to Imidazobenzoxepin Compound 3 | | Descriptor: | 10-(3-methyl-3-oxidanyl-but-1-ynyl)-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Smith, M.A, McEwan, P, Hymowitz, S.G. | | Deposit date: | 2016-09-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
J. Med. Chem., 60, 2017
|
|
6ARD
 
 | |
6B73
 
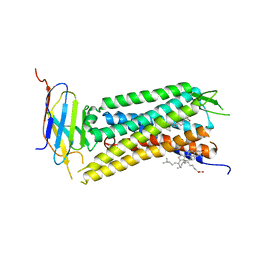 | | Crystal Structure of a nanobody-stabilized active state of the kappa-opioid receptor | | Descriptor: | CHOLESTEROL, N-[(5alpha,6beta)-17-(cyclopropylmethyl)-3-hydroxy-7,8-didehydro-4,5-epoxymorphinan-6-yl]-3-iodobenzamide, Nanobody, ... | | Authors: | Che, T, Majumdar, S, Zaidi, S.A, Kormos, C, McCorvy, J.D, Wang, S, Mosier, P.D, Uprety, R, Vardy, E, Krumm, B.E, Han, G.W, Lee, M.Y, Pardon, E, Steyaert, J, Huang, X.P, Strachan, R.T, Tribo, A.R, Pasternak, G.W, Carroll, I.F, Stevens, R.C, Cherezov, V, Katritch, V, Wacker, D, Roth, B.L. | | Deposit date: | 2017-10-03 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Nanobody-Stabilized Active State of the Kappa Opioid Receptor.
Cell, 172, 2018
|
|
3D25
 
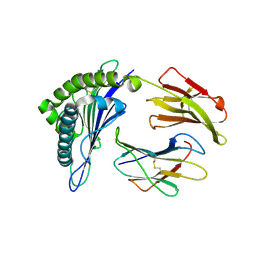 | | Crystal structure of HA-1 minor histocompatibility antigen bound to human class I MHC HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Nicholls, S, Piper, K.P, Mohammed, F, Dafforn, T.R, Tenzer, S, Salim, M, Mahendra, P, Craddock, C, van Endert, P, Schild, H, Cobbold, M, Engelhard, V.H, Moss, P.A.H, Willcox, B.E. | | Deposit date: | 2008-05-07 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Secondary anchor polymorphism in the HA-1 minor histocompatibility antigen critically affects MHC stability and TCR recognition
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5UE3
 
 | | proMMP-9desFnII | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Matrix metalloproteinase-9, ... | | Authors: | Alexander, R.S, Spurlino, J, Milligan, C. | | Deposit date: | 2016-12-29 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Discovery of a highly selective chemical inhibitor of matrix metalloproteinase-9 (MMP-9) that allosterically inhibits zymogen activation.
J. Biol. Chem., 292, 2017
|
|
5KQY
 
 | | Protease E35D-DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease E35D-DRV | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5KR1
 
 | | Protease PR5-DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease PR5-DRV | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
3Q2X
 
 | |
5KQX
 
 | | Protease E35D-SQV | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease E35D-SQV | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
3PZZ
 
 | |
6VLT
 
 | | Crystal Structure of Human P450 2C9*2 Genetic Variant in Complex with Losartan | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2C9, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shah, M.B. | | Deposit date: | 2020-01-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structure of Cytochrome P450 2C9*2 in Complex with Losartan: Insights into the Effect of Genetic Polymorphism.
Mol.Pharmacol., 98, 2020
|
|
6W3N
 
 | | APE1 exonuclease substrate complex D148E | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(C7R))-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
