8HZJ
 
 | | A new fluorescent RNA aptamer bound with N571 | | Descriptor: | (5~{Z})-5-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8HZK
 
 | | A new fluorescent RNA aptamer bound with N, iridium hexammine soak | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, DI(HYDROXYETHYL)ETHER, IRIDIUM HEXAMMINE ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8HZF
 
 | | A new fluorescent RNA aptamer bound with N565 | | Descriptor: | (5~{Z})-5-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]methylidene]-2-[(~{E})-2-(4-hydroxyphenyl)ethenyl]-3-methyl-imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8HZL
 
 | | A new fluorescent RNA aptamer_III bound with N | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, MAGNESIUM ION, RNA (84-MER) | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
6V9D
 
 | |
6V9B
 
 | |
6EO6
 
 | | X-ray structure of the complex between human alpha-thrombin and modified 15-mer DNA aptamer containing 5-(3-(2-(1H-indol-3-yl)acetamide-N-yl)-1-propen-1-yl)-2'-deoxyuridine residue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GA63A - TBA MODIFIED APTAMER, ... | | Authors: | Dolot, R.M, Nawrot, B, Yang, X. | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity.
Nucleic Acids Res., 46, 2018
|
|
6EO7
 
 | | X-ray structure of the complex between human alpha-thrombin and modified 15-mer DNA aptamer containing 5-(3-(acetamide-N-yl)-1-propen-1-yl)-2'-deoxyuridine residue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GA68B2 - MODIFIED HUMAN THROMBIN BINDING APTAMER, ... | | Authors: | Dolot, R.M, Nawrot, B, Yang, X. | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity.
Nucleic Acids Res., 46, 2018
|
|
1DX1
 
 | | BOVINE PRION PROTEIN RESIDUES 23-230 | | Descriptor: | PRION PROTEIN | | Authors: | Lopez Garcia, F, Zahn, R, Riek, R, Billeter, M, Wuthrich, K. | | Deposit date: | 1999-12-15 | | Release date: | 2000-07-20 | | Last modified: | 2015-08-05 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Bovine Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DX0
 
 | | BOVINE PRION PROTEIN RESIDUES 23-230 | | Descriptor: | PRION PROTEIN | | Authors: | Lopez-Garcia, F, Zahn, R, Riek, R, Billeter, M, Wuthrich, K. | | Deposit date: | 1999-12-15 | | Release date: | 2000-07-20 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Bovine Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EG6
 
 | | CRYSTAL STRUCTURE ANALYSIS OF D(CG(5-BRU)ACG) COMPLEXES TO A PHENAZINE | | Descriptor: | 5'-D(*CP*GP*(BRO)UP*AP*CP*G)-3', 9-BROMO-PHENAZINE-1-CARBOXYLIC ACID (2-DIMETHYLAMINO-ETHYL)-AMIDE, BROMIDE ION, ... | | Authors: | Cardin, C.J, Denny, W.A, Hobbs, J.R, Thorpe, J.H. | | Deposit date: | 2000-02-14 | | Release date: | 2001-01-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Guanine specific binding at a DNA junction formed by d[CG(5-BrU)ACG](2) with a topoisomerase poison in the presence of Co(2+) ions.
Biochemistry, 39, 2000
|
|
7WU8
 
 | |
8PYW
 
 | | Crystal structure of the human Nucleoside-diphosphate kinase B domain bound to compound diphosphate form of AT-9052-Sp. | | Descriptor: | GLYCEROL, Nucleoside diphosphate kinase B, [[(2R,3R,4R,5R)-5-(2-azanyl-6-oxidanylidene-1H-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methoxy-sulfanyl-phosphoryl] dihydrogen phosphate | | Authors: | Feracci, M, Chazot, A. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | An exonuclease-resistant chain-terminating nucleotide analogue targeting the SARS-CoV-2 replicase complex.
Nucleic Acids Res., 52, 2024
|
|
8UFM
 
 | | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2 | | Descriptor: | ACETATE ION, FORMIC ACID, Papain-like protease nsp3, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2
To Be Published
|
|
8UFL
 
 | | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus
To Be Published
|
|
8PO8
 
 | | Structure of Escherichia coli HrpA in complex with ADP and oligonucleotide poly(dC)11 forming an i-motif | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpA, DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
8PO7
 
 | | Structure of Escherichia coli HrpA in complex with ADP and dinucleotide dCdC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpA, DNA (5'-D(P*CP*C)-3'), ... | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
8PO6
 
 | | Structure of Escherichia coli HrpA apo form | | Descriptor: | ATP-dependent RNA helicase HrpA, PHOSPHATE ION | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
8U5P
 
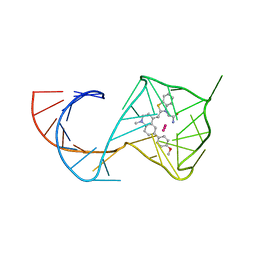 | | Structure of Mango II aptamer bound to T01-6A-B | | Descriptor: | 3-[2,16-dioxo-20-(2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)-6,9,12-trioxa-3,15-diazaicosan-1-yl]-2-{(E)-[6-(4-methoxyphenyl)-1-methylquinolin-4(1H)-ylidene]methyl}-1,3-benzothiazol-3-ium, Mango II, POTASSIUM ION | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-12 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Mango II aptamer bound to T01-6A-B
To Be Published
|
|
8U60
 
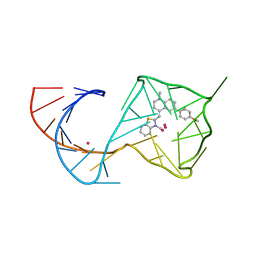 | |
8U5R
 
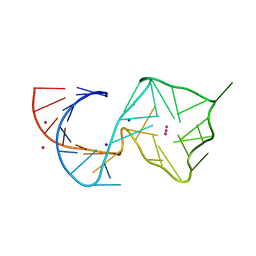 | |
8U5T
 
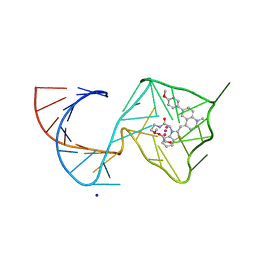 | | Structure of Mango II variant aptamer bound to T01-6A-B | | Descriptor: | 3-{2,16-dioxo-20-[(3aS,4R,6aS)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-2-{(E)-[6-(4-methoxyphenyl)-1-methylquinolin-4(1H)-ylidene]methyl}-1,3-benzothiazol-3-ium, Mango II variant, POTASSIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-12 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Mango II variant aptamer bound to T01-6A-B
To Be Published
|
|
8U5J
 
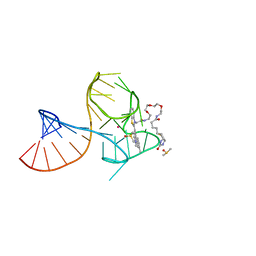 | | Structure of Mango III variant aptamer bound to T01-07M-B | | Descriptor: | 2-[(E)-(1,7-dimethylquinolin-4(1H)-ylidene)methyl]-3-{2,16-dioxo-20-[(3aR,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-1,3-benzothiazol-3-ium, DIMETHYL SULFOXIDE, Mango III variant, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-12 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Mango III variant aptamer bound to T01-07M-B
To Be Published
|
|
8U5K
 
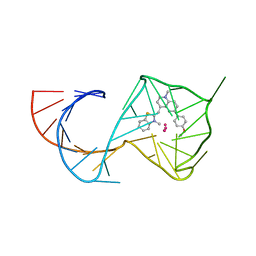 | |
8U5Z
 
 | | Structure of Mango II variant aptamer bound to T01-7M-B | | Descriptor: | 2-[(E)-(1,7-dimethylquinolin-4(1H)-ylidene)methyl]-3-{2,16-dioxo-20-[(3aR,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-1,3-benzothiazol-3-ium, Mango II variant, POTASSIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-13 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Mango II variant aptamer bound to T01-7M-B
To Be Published
|
|
