2LZP
 
 | |
4KW9
 
 | |
4KW8
 
 | |
4KW4
 
 | |
2LZQ
 
 | |
4N0H
 
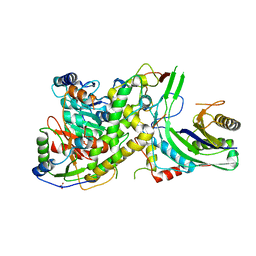 | | Crystal structure of S. cerevisiae mitochondrial GatFAB | | Descriptor: | Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial, Glutamyl-tRNA(Gln) amidotransferase subunit B, ... | | Authors: | Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-10-02 | | Release date: | 2014-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases
Nucleic Acids Res., 42, 2014
|
|
4N0I
 
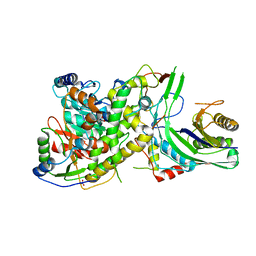 | | Crystal structure of S. cerevisiae mitochondrial GatFAB in complex with glutamine | | Descriptor: | GLUTAMINE, Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial, ... | | Authors: | Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-10-02 | | Release date: | 2014-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases
Nucleic Acids Res., 42, 2014
|
|
4JZK
 
 | |
4KA9
 
 | |
2MKB
 
 | |
4OGS
 
 | |
4P7H
 
 | |
4Q9W
 
 | |
4Q9X
 
 | | mTFP* PdCl2 soak | | Descriptor: | CARBONATE ION, CHLORIDE ION, GFP-like fluorescent chromoprotein cFP484, ... | | Authors: | Fischer, J, Quitterer, F, Groll, M, Eppinger, J. | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | mTFP*: An imperishable and versatile protein host for anchoring diverse ligands and organocatalysts
To be Published
|
|
4OWL
 
 | |
4OWJ
 
 | |
4CQH
 
 | | Structure of Infrared Fluorescent Protein IFP2.0 | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, BACTERIOPHYTOCHROME, SODIUM ION | | Authors: | Lafaye, C, Yu, D, Noirclerc-Savoye, M, Shu, X, Royant, A. | | Deposit date: | 2014-02-17 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | An Improved Monomeric Infrared Fluorescent Protein for Neuronal and Tumour Brain Imaging.
Nat.Commun., 5, 2014
|
|
4OWK
 
 | |
3J6Y
 
 | | S. cerevisiae 80S ribosome bound with Taura syndrome virus (TSV) IRES, 2 degree rotation (Class I) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Koh, C.S, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-04-16 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3J6X
 
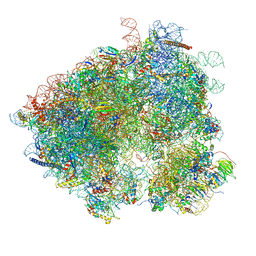 | | S. cerevisiae 80S ribosome bound with Taura syndrome virus (TSV) IRES, 5 degree rotation (Class II) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Koh, C.S, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-04-16 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4KZE
 
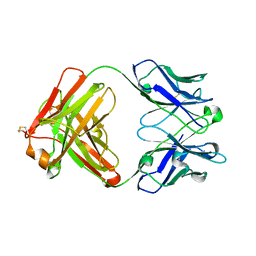 | | Crystal structure of an RNA aptamer in complex with Fab | | Descriptor: | BL3-6 Fab antibody, heavy chain, light chain, ... | | Authors: | Huang, H, Suslov, N.B, Li, N, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2013-05-29 | | Release date: | 2014-06-18 | | Last modified: | 2014-07-30 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
4PL0
 
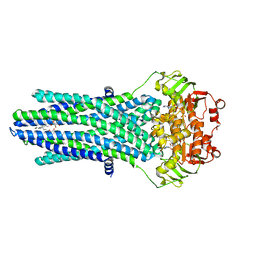 | |
4KZD
 
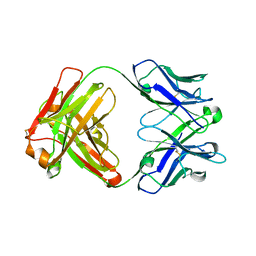 | | Crystal structure of an RNA aptamer in complex with fluorophore and Fab | | Descriptor: | 4-(3,5-difluoro-4-hydroxybenzyl)-1,2-dimethyl-1H-imidazol-5-ol, BL3-6 Fab antibody, heavy chain, ... | | Authors: | Huang, H, Suslov, N.B, Li, N, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2013-05-29 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
4Q9Q
 
 | | Crystal structure of an RNA aptamer bound to bromo-ligand analog in complex with Fab | | Descriptor: | (5Z)-5-(3-bromobenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, Fab BL3-6, HEAVY CHAIN, ... | | Authors: | Huang, H, Suslov, N.B, Li, N.-S, Shelke, S.A, Evans, M.E, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-18 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
4Q9R
 
 | | Crystal structure of an RNA aptamer bound to trifluoroethyl-ligand analog in complex with Fab | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Fab BL3-6, HEAVY CHAIN, ... | | Authors: | Huang, H, Suslov, N.B, Li, N.-S, Shelke, S.A, Evans, M.E, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-18 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
