6OQA
 
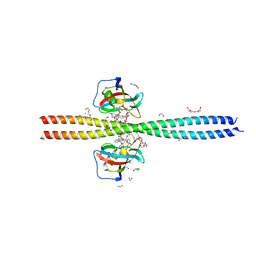 | | Crystal structure of CEP250 bound to FKBP12 in the presence of FK506-like novel natural product | | Descriptor: | (3R,4E,7E,10R,11S,12R,13S,16R,17R,24aS)-11,17-dihydroxy-10,12,16-trimethyl-3-[(2R)-1-phenylbutan-2-yl]-6,9,10,11,12,13,14,15,16,17,22,23,24,24a-tetradecahydro-3H-13,17-epoxypyrido[2,1-c][1,4]oxazacyclohenicosine-1,18,19(21H)-trione, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Lee, S.-J, Shigdel, U.K, Townson, S.A, Verdine, G.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genomic discovery of an evolutionarily programmed modality for small-molecule targeting of an intractable protein surface.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P2J
 
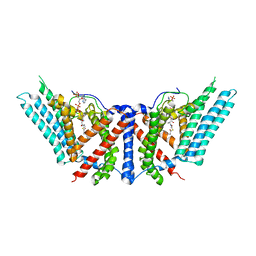 | | Dimeric structure of ACAT1 | | Descriptor: | S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name), Sterol O-acyltransferase 1 | | Authors: | Yan, N, Qian, H.W. | | Deposit date: | 2019-05-21 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for catalysis and substrate specificity of human ACAT1.
Nature, 581, 2020
|
|
7RKP
 
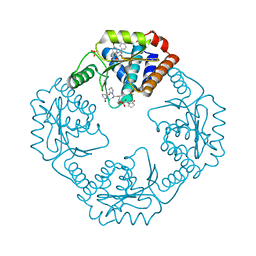 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with cyclic compound SJ001034733 | | Descriptor: | (5R)-5-hydroxy-16,21-dioxa-3,8,28-triazatetracyclo[20.3.1.1~2,6~.1~11,15~]octacosa-1(26),2,6(28),11(27),12,14,22,24-octaene-4,7-dione, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
3SU4
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease,NS4A protein, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SU3
 
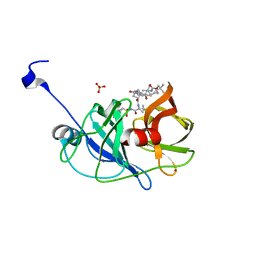 | | Crystal structure of NS3/4A protease in complex with vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SUG
 
 | | Crystal structure of NS3/4A protease variant A156T in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
6PIV
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-7 (NR03-77) | | Descriptor: | (1R,3r,5S)-bicyclo[3.1.0]hexan-3-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6PJ0
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-5 (NR01-97) | | Descriptor: | 1,2-ETHANEDIOL, 1-ethylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4 Aprotease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6P2P
 
 | | Tetrameric structure of ACAT1 | | Descriptor: | S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name), Sterol O-acyltransferase 1 | | Authors: | Yan, N, Qian, H.W. | | Deposit date: | 2019-05-21 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for catalysis and substrate specificity of human ACAT1.
Nature, 581, 2020
|
|
3SUE
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
7RTK
 
 | | Structure of the (NIAU)2 complex with N-terminal mutation of ISCU2 Y35D at 2.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ... | | Authors: | Boniecki, M.T, Cygler, M. | | Deposit date: | 2021-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The essential function of ISCU2 and its conserved N-terminus in Fe/S cluster biogenesis
To Be Published
|
|
6PIW
 
 | |
6PJ1
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-4(AJ-74) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
3SUF
 
 | | Crystal structure of NS3/4A protease variant D168A in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SU2
 
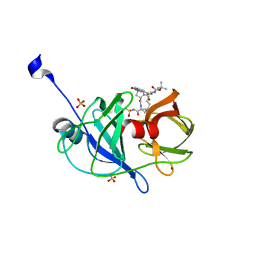 | | Crystal structure of NS3/4A protease variant A156T in complex with danoprevir | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, Genome polyprotein, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
6PIZ
 
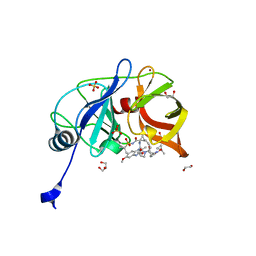 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-1 (NR02-24) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclopropyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, GLYCEROL, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
3V4X
 
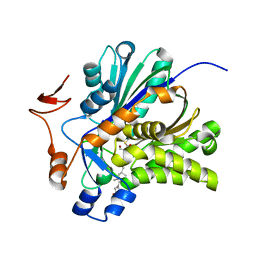 | | The Biochemical and Structural Basis for Inhibition of Enterococcus faecalis HMG-CoA Synthase, mvaS, by Hymeglusin | | Descriptor: | (7R,12R,13R)-13-formyl-12,14-dihydroxy-3,5,7-trimethyltetradeca-2,4-dienoic acid, HMG-CoA synthase | | Authors: | Skaff, D.A, Ramyar, K.X, McWhorter, W.J, Geisbrecht, B.V, Miziorko, H.M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Biochemical and structural basis for inhibition of Enterococcus faecalis hydroxymethylglutaryl-CoA synthase, mvaS, by hymeglusin.
Biochemistry, 51, 2012
|
|
6PJ2
 
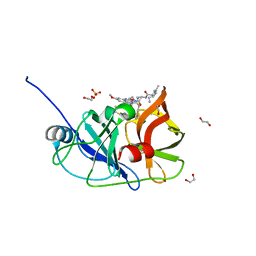 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-P5-4 (AJ-65) | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(N-acetyl-L-isoleucyl)amino]-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-N-[(1-methylcyclo propyl)sulfonyl]-5,16-dioxo-1,2,3,6,7,8,9,10,11,13a,14,15,16,16a-tetradecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacycl opentadecine-14a(5H)-carboxamide, 1,2-ETHANEDIOL, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6WIH
 
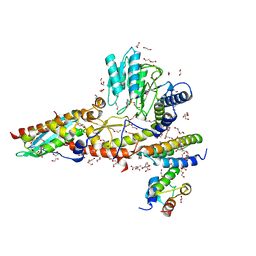 | | N-terminal mutation of ISCU2 (L35H36) traps Nfs1 Cys loop in the active site of ISCU2 without metal present. Structure of human mitochondrial complex Nfs1-ISCU2(L35H36)-ISD11 with E.coli ACP1 at 1.9 A resolution (NIAU)2. | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, Acyl carrier protein, ... | | Authors: | Boniecki, M.T, Cygler, M. | | Deposit date: | 2020-04-09 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The essential function of ISCU2 and its conserved N-terminus in Fe/S cluster biogenesis
To Be Published
|
|
6PIU
 
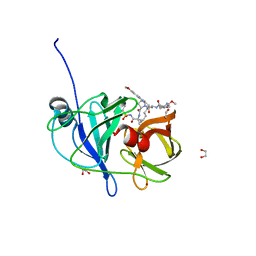 | |
6PIY
 
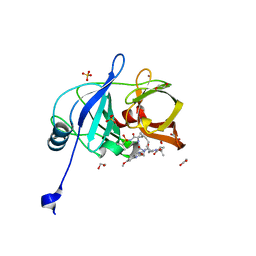 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-2 (NR02-61) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclobutyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6PIX
 
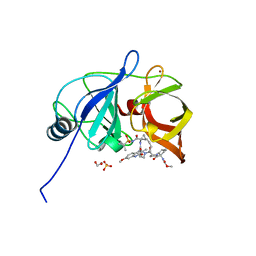 | |
7QGF
 
 | | Cubic Insulin SAD phasing at 14.2 keV | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Nanao, M.H, Basu, S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.203 Å) | | Cite: | ID23-2: an automated and high-performance microfocus beamline for macromolecular crystallography at the ESRF.
J.Synchrotron Radiat., 29, 2022
|
|
6PYD
 
 | | Structure of 3E9 antibody Fab bound to marinobufagenin | | Descriptor: | (3beta,5beta,14alpha,15beta)-3,5-dihydroxy-14,15-epoxybufa-20,22-dienolide, 3E9 anti-marinobufagenin antibody Fab heavy chain, recloned with human IgG4 C region, ... | | Authors: | Franklin, M.C, Macdonald, L.E, McWhirter, J, Murphy, A.J. | | Deposit date: | 2019-07-29 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kappa-on-Heavy (KoH) bodies are a distinct class of fully-human antibody-like therapeutic agents with antigen-binding properties.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3T6E
 
 | | Crystal Structure of the Reaction Centre from Blastochloris viridis strain DSM 133 (ATCC 19567) substrain-94 | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | New insights into the structure of the reaction centre from Blastochloris viridis: evolution in the laboratory.
Biochem.J., 442, 2012
|
|
